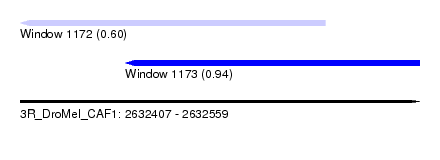
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,632,407 – 2,632,559 |
| Length | 152 |
| Max. P | 0.942009 |

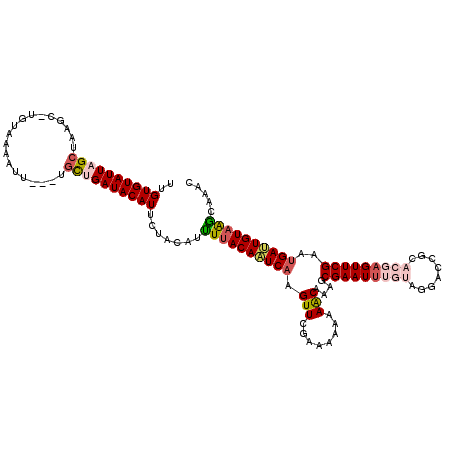
| Location | 2,632,407 – 2,632,523 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -13.63 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2632407 116 - 27905053 UUCUACAUUUUACAAUCAAGUUCGA----AAACAAAGCGAAUAUGUCGGACCGCACGAGUUCGAAUGAUUGUAAGCAAACUCGAAAAUGUCAUUAUGCAGUCGAACGAAAUUAAUAUUUA (((.....((((((((((.(((((.----........)))))...(((((((....).)))))).)))))))))).....((((...(((......))).))))..)))........... ( -23.90) >DroSec_CAF1 32560 115 - 1 UUCUACAUUUUACAAUCAAGUUCGAAUAAAAACGAAGCGAAUUUGUAGGACCGCACGAGUUCGAAUGAUUGUAAGCAAGCUUGAAAAUGUCAUUAUGCAUUCGAACGAAAAUAAU----- ...................((((((((..........(((((((((........)))))))))((((((..((((....)))).....))))))....)))))))).........----- ( -24.70) >DroSim_CAF1 20788 120 - 1 UUCUACAUUUUACAAUCAAGUUCGAAUAAAAACGAAGCGAAUUUGUAGGACCGCACGAGUUCGAAUGAUUGUAAGCAAGCUGGAAAAUGUCAUUAUGCAUUCGAACGAAAUUAAUAUUUA ((((((..((((((((((..((((........)))).(((((((((........)))))))))..))))))))))...).)))))(((((......)))))................... ( -23.10) >DroEre_CAF1 22113 112 - 1 UUCUACAUUUUACAAUCUAGUUCGAAAGAAAACAAGGCGAAUUUGUAGG--------AGUUCGAAUGAAUGUUAACAAUCUCGAACAAGUCGUAAUGCAUUCGAACCAAAUUAAUAUUUA (((((((..((.........(((....))).........))..))))))--------)(((((((((..((....))((..(((.....)))..)).))))))))).............. ( -23.17) >DroYak_CAF1 14416 109 - 1 UUCUACAUCUUACAUUCAAGUUCGAAAGAAAGCAAAGCGAAUUCGUAGG--------AGUUCGAAUGAAUGUAGGCAAACUCGACAAAA--AUGAUGAGUUCGAACACAAUU-AUAUUUA ........((((((((((..(((....))).......(((((((....)--------))))))..))))))))))(.((((((.((...--.)).)))))).).........-....... ( -27.00) >consensus UUCUACAUUUUACAAUCAAGUUCGAAAAAAAACAAAGCGAAUUUGUAGGACCGCACGAGUUCGAAUGAUUGUAAGCAAACUCGAAAAUGUCAUUAUGCAUUCGAACGAAAUUAAUAUUUA ............(...((((((((.............))))))))...).........(((((((((...((((..................)))).))))))))).............. (-13.63 = -14.27 + 0.64)

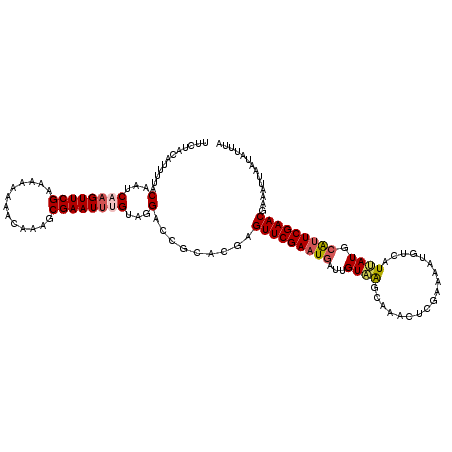
| Location | 2,632,447 – 2,632,559 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -20.71 |
| Energy contribution | -22.27 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2632447 112 - 27905053 UUGUGUAUUAGCUAAGU-UGUAAAAUU---UUCUGAUACAUUCUACAUUUUACAAUCAAGUUCGA----AAACAAAGCGAAUAUGUCGGACCGCACGAGUUCGAAUGAUUGUAAGCAAAC ..(((((((((..((((-(....))))---).))))))))).......((((((((((.(((((.----........)))))...(((((((....).)))))).))))))))))..... ( -28.30) >DroSec_CAF1 32595 117 - 1 UUGUGUAUUAGCUAAGCGUGUAAAAUU---UGCUGAUACAUUCUACAUUUUACAAUCAAGUUCGAAUAAAAACGAAGCGAAUUUGUAGGACCGCACGAGUUCGAAUGAUUGUAAGCAAGC ..((((((((((...............---.)))))))))).......((((((((((..((((........)))).(((((((((........)))))))))..))))))))))..... ( -30.79) >DroSim_CAF1 20828 117 - 1 UUGUGUAUUAGCUAAGCGUGUAAAAUU---UGCUGAUACAUUCUACAUUUUACAAUCAAGUUCGAAUAAAAACGAAGCGAAUUUGUAGGACCGCACGAGUUCGAAUGAUUGUAAGCAAGC ..((((((((((...............---.)))))))))).......((((((((((..((((........)))).(((((((((........)))))))))..))))))))))..... ( -30.79) >DroEre_CAF1 22153 108 - 1 UUGUGUAUUAGCUAAGC-CCUGGAAUU---CGCAGAUACAUUCUACAUUUUACAAUCUAGUUCGAAAGAAAACAAGGCGAAUUUGUAGG--------AGUUCGAAUGAAUGUUAACAAUC ((((.((((.....(((-(((((((((---((((((.....)))................(((....)))......)))))))).))))--------.)))......))))...)))).. ( -21.00) >DroYak_CAF1 14453 111 - 1 UUGUGUAUUAGCUAAGC-UUUGUAUUUGUGUGUAGAUACAUUCUACAUCUUACAUUCAAGUUCGAAAGAAAGCAAAGCGAAUUCGUAGG--------AGUUCGAAUGAAUGUAGGCAAAC ((((.(((...(...((-(((((...((((((((((.....)))))))...)))......(((....))).)))))))((((((....)--------)))))....)...))).)))).. ( -30.80) >consensus UUGUGUAUUAGCUAAGC_UGUAAAAUU___UGCUGAUACAUUCUACAUUUUACAAUCAAGUUCGAAAAAAAACAAAGCGAAUUUGUAGGACCGCACGAGUUCGAAUGAUUGUAAGCAAAC ..((((((((((...................)))))))))).......((((((((((.(((........)))....(((((((((........)))))))))..))))))))))..... (-20.71 = -22.27 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:47 2006