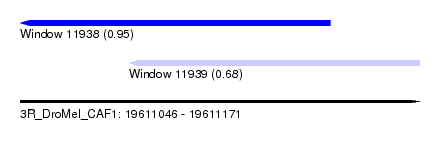
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,611,046 – 19,611,171 |
| Length | 125 |
| Max. P | 0.951396 |

| Location | 19,611,046 – 19,611,143 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 71.27 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -8.62 |
| Energy contribution | -9.80 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
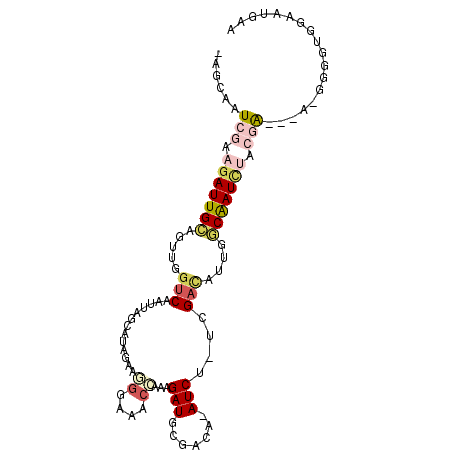
>3R_DroMel_CAF1 19611046 97 - 27905053 -AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA-AUCU-UCGACAUUGGCAAUCUUCGAA--ACGGGGUGGAAUGAA -.....(((((((((((.....(((.....((((....((....))....))))((..-....-)))))....))))))))))).--............... ( -30.90) >DroVir_CAF1 18467 89 - 1 CAGCUUUCCGCGAUUGGAAAAUGCCAAUUUACAAU---UUACAACAAAUGAUGCGACA-AUCUCUUGAUAUUGUCGAUUUAUAG---------AGCAAUUAA ..(((((....((((((......))))))......---.........((((..(((((-((........)))))))..))))))---------)))...... ( -16.90) >DroSec_CAF1 17526 97 - 1 -AGCAAUCGAAAAUUGCAGUUGGUCAAUUAGCAUAGAAUGGAAACCAAAGAUGCAACA-AUCU-UCGACAUUGGCAAUCUUCGAA--GCGGGGUGGAAUGAA -.((..(((((.(((((.(((((.......((((.....(....).....))))....-....-)))))....))))).))))).--))............. ( -21.06) >DroEre_CAF1 18070 97 - 1 -AGCAAUCGAAGAUUGCAGUUGAUCAAUUAGCAUAUAAGGGAAACCAAAGAUGCGACAAAUCU-CCGACAUUGGCAAUCUGCGA---AGGGGGUGGAAUGAA -.((((((...))))))......(((....((((....((....))....)))).....((((-((....((.((.....)).)---)))))))....))). ( -24.60) >DroYak_CAF1 17680 99 - 1 -AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA-AUCU-CCGACAUUGGCAAUCUGCGAAGAAGGGGGUGGAAUGAA -.....(((.(((((((.(((((.......((((....((....))....))))....-....-)))))....))))))).))).................. ( -28.46) >DroMoj_CAF1 18631 83 - 1 UUGCUUUCCACGAUUGGAAAAUGUCAAUUUACAA-------CA--AAAAGAUACGACA-AUCUUUUGACAUUGGCAAUUUAUAG---------AGCAAUCAA (((((((....(((((...((((((((.......-------..--..(((((......-)))))))))))))..)))))...))---------))))).... ( -17.20) >consensus _AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA_AUCU_UCGACAUUGGCAAUCUACGA___A_GGGGUGGAAUGAA ......(((.(((((((.....(((.............((....))...(((.......)))....)))....))))))).))).................. ( -8.62 = -9.80 + 1.18)


| Location | 19,611,080 – 19,611,171 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -12.19 |
| Energy contribution | -13.98 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19611080 91 - 27905053 AUUUAUGUUUAAUUA--AGCAAUUUCCGAG--AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA-AUCU-UC .((((((((..((((--((((((((.(((.--.....))).)))))))..))))).....)))))))).((....)).(((((......-))))-). ( -22.40) >DroSec_CAF1 17560 91 - 1 AUUUAUGUUUAAUUA--AGCAAUUUCCGAG--AGCAAUCGAAAAUUGCAGUUGGUCAAUUAGCAUAGAAUGGAAACCAAAGAUGCAACA-AUCU-UC .....((((......--.(((((((.(((.--.....))).))))))).............((((.....(....).....))))))))-....-.. ( -17.90) >DroSim_CAF1 17870 87 - 1 AUUUAUGUUUAAUUA--AGCAAUUUCCGAG--AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA-AU----- .....((((......--.(((((((.(((.--.....))).))))))).............((((....((....))....))))))))-..----- ( -21.90) >DroEre_CAF1 18103 92 - 1 AUUUAUGUUUAAUUA--AGCAAUUUCCGAG--AGCAAUCGAAGAUUGCAGUUGAUCAAUUAGCAUAUAAGGGAAACCAAAGAUGCGACAAAUCU-CC .....((.((((((.--.(((((((.(((.--.....))).))))))))))))).))....((((....((....))....)))).........-.. ( -22.10) >DroYak_CAF1 17716 91 - 1 AUUUAUGUUUAAUUA--AGCAAUUUCCGAG--AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA-AUCU-CC .....((((......--.(((((((.(((.--.....))).))))))).............((((....((....))....))))))))-....-.. ( -21.90) >DroMoj_CAF1 18658 80 - 1 AUUUAUGCUUAAUUACAAGCAAU-------UUUGCUUUCCACGAUUGGAAAAUGUCAAUUUACAA-------CA--AAAAGAUACGACA-AUCUUUU .....(((((......)))))..-------.....((((((....))))))..............-------..--(((((((......-))))))) ( -11.80) >consensus AUUUAUGUUUAAUUA__AGCAAUUUCCGAG__AGCAAUCGAAGAUUGCAGUUGGUCAAUUAGCAUAGAAGGGAAACCAAAGAUGCGACA_AUCU_UC .....((((.........(((((((.(((........))).))))))).............((((....((....))....))))))))........ (-12.19 = -13.98 + 1.79)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:38 2006