| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
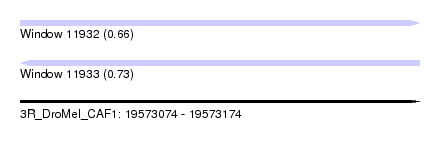
| Location | 19,573,074 – 19,573,174 |
| Length | 100 |
| Max. P | 0.730738 |

| Location | 19,573,074 – 19,573,174 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -22.40 |
| Energy contribution | -23.85 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
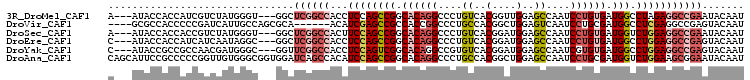
>3R_DroMel_CAF1 19573074 100 + 27905053 A---AUACCACCAUCGUCUAUGGGU---GGCUCGGCCACCUCCAGCCGGCACAGGCCCUGUCACGGUUGGAGCCAAUCCUGUGAUGGCCUAGAGGCCGAAUACAAU .---...(((((..........)))---)).((((((...((((((((..((((...))))..))))))))((((.((....)))))).....))))))....... ( -40.50) >DroVir_CAF1 222 96 + 1 ----GCGCCACCCCCGAUCAUUGCCAGCGCA------ACAUCGAGCCGCCACCGGCCCUGCCACGGCUGGAGUCAAUCCUGCGAUGGCCUCGAGGCCGAGUACAAU ----(((.....(.((((..((((....)))------).)))).).))).((((((((.((((..((.(((.....))).))..))))...).))))).))..... ( -29.10) >DroSec_CAF1 256 100 + 1 A---AUACCACCACCGUCUAUGGGU---GGCUCGGCCACUUCCAGCCGGCACAGGCCCUGUCACGGAUGGAGCCAAUCCUGUGAUGGUCUGGAGGCCGAAUACAAU .---......(((((.......)))---)).((((((...((((((((((....)))..((((((((((....).)))).))))))).)))))))))))....... ( -42.00) >DroEre_CAF1 246 100 + 1 C---AUACCACCAUCAUCAAUAGGC---GGCUCGGCCACCUCCAGCCGGCACAGGCCCUGUCACGGAUGGAGCCAAUCCUGUGAUGGCCUGGAGGCCGAGUACAAU .---...((.((..........)).---))(((((((...((((((((((....)))..((((((((((....).)))).))))))).))))))))))))...... ( -39.90) >DroYak_CAF1 258 100 + 1 C---AUACCGCCGCCAACGAUGGGC---GGUUCGGCCACCUCCAGUCGGCACAGGCCGUGUCACGGAUGGAGCCAAUCGUGUGAUGGCCUGGAGGCCGAGUACAAU .---.(((.((((((.......)))---)))((((((....((....))..((((((((..(((((.((....)).)))))..))))))))..))))))))).... ( -46.00) >DroAna_CAF1 249 106 + 1 CAGCAUUCCGCCCCCGGUUGUGGGCGGUGGAUCAGCCACAUCCAGCCGGCACAGGCCCUGCCACGGCUGGAGCCAAUCCUGCGAUGGUCUGGAAGCGGAAUACAAU ..(.(((((((..((((.....(((.((((.....)))).((((((((.....(((...))).)))))))))))...((......)).))))..))))))).)... ( -45.20) >consensus C___AUACCACCACCGUCUAUGGGC___GGCUCGGCCACCUCCAGCCGGCACAGGCCCUGUCACGGAUGGAGCCAAUCCUGUGAUGGCCUGGAGGCCGAAUACAAU ...............................((((((...((((((((.((((((....((..(....)..))....)))))).))).)))))))))))....... (-22.40 = -23.85 + 1.45)

| Location | 19,573,074 – 19,573,174 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
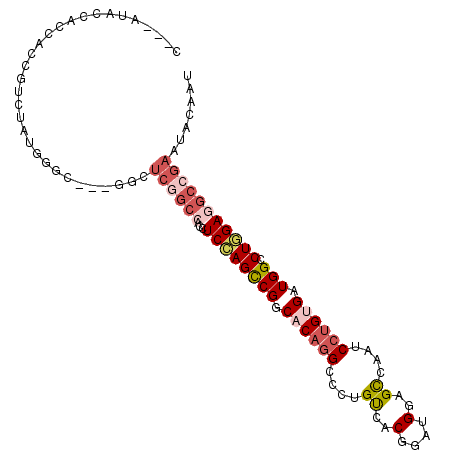
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -25.37 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
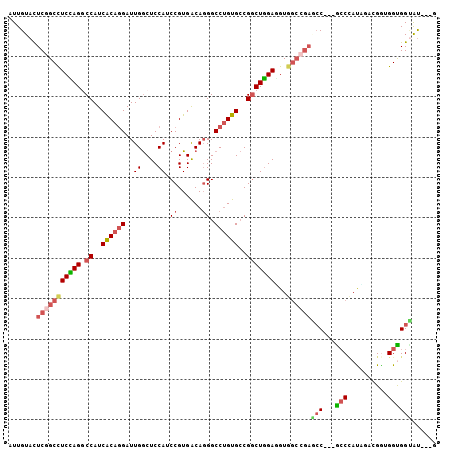
>3R_DroMel_CAF1 19573074 100 - 27905053 AUUGUAUUCGGCCUCUAGGCCAUCACAGGAUUGGCUCCAACCGUGACAGGGCCUGUGCCGGCUGGAGGUGGCCGAGCC---ACCCAUAGACGAUGGUGGUAU---U .......(((((((((.((((..((((((.(((....)))((......)).))))))..))))..))).))))))(((---(((.((.....))))))))..---. ( -41.90) >DroVir_CAF1 222 96 - 1 AUUGUACUCGGCCUCGAGGCCAUCGCAGGAUUGACUCCAGCCGUGGCAGGGCCGGUGGCGGCUCGAUGU------UGCGCUGGCAAUGAUCGGGGGUGGCGC---- ......((((....))))((((((.(..(((((...(((((.(..(((((((((....))))))..)))------..))))))...))))).).))))))..---- ( -43.60) >DroSec_CAF1 256 100 - 1 AUUGUAUUCGGCCUCCAGACCAUCACAGGAUUGGCUCCAUCCGUGACAGGGCCUGUGCCGGCUGGAAGUGGCCGAGCC---ACCCAUAGACGGUGGUGGUAU---U .......(((((((((((.((.((((.((((.(....)))))))))...(((....))))))))))...))))))(((---(((.......)))))).....---. ( -42.30) >DroEre_CAF1 246 100 - 1 AUUGUACUCGGCCUCCAGGCCAUCACAGGAUUGGCUCCAUCCGUGACAGGGCCUGUGCCGGCUGGAGGUGGCCGAGCC---GCCUAUUGAUGAUGGUGGUAU---G ......(((((((..((((((.((((.((((.(....)))))))))...)))))).(((.......))))))))))((---(((..........)))))...---. ( -44.20) >DroYak_CAF1 258 100 - 1 AUUGUACUCGGCCUCCAGGCCAUCACACGAUUGGCUCCAUCCGUGACACGGCCUGUGCCGACUGGAGGUGGCCGAACC---GCCCAUCGUUGGCGGCGGUAU---G ...(((((..(((..((((((....((((..((....))..))))....)))))).((((((..(.(((((.....))---))))...))))))))))))))---. ( -43.50) >DroAna_CAF1 249 106 - 1 AUUGUAUUCCGCUUCCAGACCAUCGCAGGAUUGGCUCCAGCCGUGGCAGGGCCUGUGCCGGCUGGAUGUGGCUGAUCCACCGCCCACAACCGGGGGCGGAAUGCUG ...(((((((((((((.(.....)...((((..((((((((((..((((...))))..))))))))....))..)))).............))))))))))))).. ( -52.10) >consensus AUUGUACUCGGCCUCCAGGCCAUCACAGGAUUGGCUCCAUCCGUGACAGGGCCUGUGCCGGCUGGAGGUGGCCGAGCC___GCCCAUAGACGGUGGUGGUAU___G .......(((((((((((.((..((((((..((....)).((......)).))))))..)))))))...))))))(((...(((..........))))))...... (-25.37 = -26.23 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:32 2006