| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,630,191 – 2,630,293 |
| Length | 102 |
| Max. P | 0.661508 |

| Location | 2,630,191 – 2,630,293 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -26.47 |
| Energy contribution | -27.97 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2630191 102 + 27905053 CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGUGGCUCUGGCCUUG-GCUGCA------CAUUGAUUUAUUUAUGCGGCCGCUCUCCUUUGU---CCCCUG-------- ...(((.(((((((((.(((..(((((....))))).))).))))))))))))...(-((((((------...(((.....))))))))))............---......-------- ( -38.70) >DroPse_CAF1 27220 116 + 1 CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGUGACUCUGGCCUUGGGCUGCACUACCACAUUGAUUUA----UGCCCGGGAUCUCUUUUGUGUGCUCCUGCCACAGGG ((((..((((..(((((..((((((((....)))))..............(..(((((((....................----.)))))))..).)))..)))))..))).)..)))). ( -39.00) >DroSim_CAF1 18526 102 + 1 CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGUGGCUCUGGCCUUG-GCUGCA------CAUUGAUUUAUUUAUGCGGCCGGACUCCUUUGU---CCCCUG-------- ...(((.(((((((((.(((..(((((....))))).))).))))))))))))...(-((((((------...(((.....))))))))))((((......))---))....-------- ( -44.30) >DroEre_CAF1 19963 102 + 1 CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGUGGCUCUGGCCUUG-GCUGCA------CAUUGAUUUAUUUAUGCGGCCGCUCUCCUUUGU---CCCCGG-------- ((.(((.(((((((((.(((..(((((....))))).))).))))))))))))...(-((((((------...(((.....))))))))))............---....))-------- ( -38.90) >DroYak_CAF1 12429 99 + 1 CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGCGGCUCUGGCCUUG-GCUGCA------CUUUGAUUUAUUUAUGCGGCCGCUC---UUUGU---CCCCUG-------- ...(((.((((((.((.(((..(((((....))))).))).)).)))))))))...(-((((((------...(((.....))))))))))....---.....---......-------- ( -34.80) >DroPer_CAF1 27333 116 + 1 CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGUGACUCUGGCCUUGGGCUGCACUACCACAUUGAUUUA----UGCCCGGGAUCUCUUUUGUGUGCUCCUGCCACACGG ...(((.((((.((((.(((..(((((....))))).))).)))).)))))))(((((((....................----.)))))))........(((((((....).)))))). ( -37.20) >consensus CCUGCCGAGGGCCACACUUGAGGUGCAUUGUUGCACAUAAUUGUGGCUCUGGCCUUG_GCUGCA______CAUUGAUUUAUUUAUGCGGCCGCUCUCCUUUGU___CCCCUG________ ...(((.(((((((((.(((..(((((....))))).))).)))))))))))).....((((((....................)))))).............................. (-26.47 = -27.97 + 1.50)

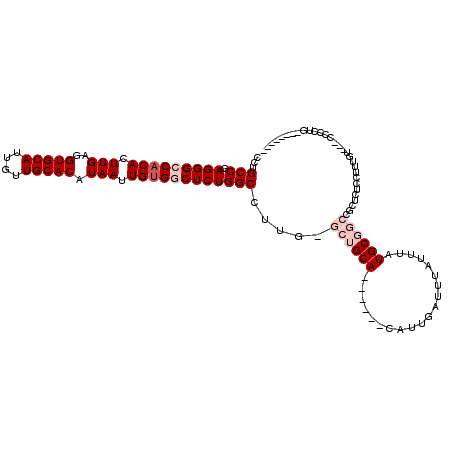
| Location | 2,630,191 – 2,630,293 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -22.94 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2630191 102 - 27905053 --------CAGGGG---ACAAAGGAGAGCGGCCGCAUAAAUAAAUCAAUG------UGCAGC-CAAGGCCAGAGCCACAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG --------..(...---.)..........(((.(((((..........))------))).))-)...(((((.((((((.....(((((....)))))......)))))).)).)))... ( -35.10) >DroPse_CAF1 27220 116 - 1 CCCUGUGGCAGGAGCACACAAAAGAGAUCCCGGGCA----UAAAUCAAUGUGGUAGUGCAGCCCAAGGCCAGAGUCACAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG .(((((.(.(((.((.((((...(((...((..(((----(......))))))..((((((((...)))....(.((((....))))).....))))))))...)))))))))).))))) ( -36.90) >DroSim_CAF1 18526 102 - 1 --------CAGGGG---ACAAAGGAGUCCGGCCGCAUAAAUAAAUCAAUG------UGCAGC-CAAGGCCAGAGCCACAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG --------....((---((......))))(((.(((((..........))------))).))-)...(((((.((((((.....(((((....)))))......)))))).)).)))... ( -39.10) >DroEre_CAF1 19963 102 - 1 --------CCGGGG---ACAAAGGAGAGCGGCCGCAUAAAUAAAUCAAUG------UGCAGC-CAAGGCCAGAGCCACAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG --------((((((---.....((.....(((.(((((..........))------))).))-)....))...((((((.....(((((....)))))......)))))))))))).... ( -37.10) >DroYak_CAF1 12429 99 - 1 --------CAGGGG---ACAAA---GAGCGGCCGCAUAAAUAAAUCAAAG------UGCAGC-CAAGGCCAGAGCCGCAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG --------..(...---.)...---....(((.((((............)------))).))-)...(((((.((((((.....(((((....)))))......)))))).)).)))... ( -33.70) >DroPer_CAF1 27333 116 - 1 CCGUGUGGCAGGAGCACACAAAAGAGAUCCCGGGCA----UAAAUCAAUGUGGUAGUGCAGCCCAAGGCCAGAGUCACAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG ..(((((.......)))))..........((((((.----...(((.....)))......))))...(((((.((((((.....(((((....)))))......)))))).)).))).)) ( -34.60) >consensus ________CAGGGG___ACAAAGGAGAGCGGCCGCAUAAAUAAAUCAAUG______UGCAGC_CAAGGCCAGAGCCACAAUUAUGUGCAACAAUGCACCUCAAGUGUGGCCCUCGGCAGG .................................(((....................)))........(((((.((((((.....(((((....)))))......)))))).)).)))... (-22.94 = -22.58 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:43 2006