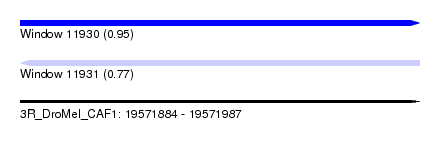
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,571,884 – 19,571,987 |
| Length | 103 |
| Max. P | 0.950364 |

| Location | 19,571,884 – 19,571,987 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -19.57 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19571884 103 + 27905053 UACACAAUG-CUUAG----CCACUUUGGUGCUGGUACUC--GCACUCGCACUCGUAAAUCCCAAUGAAUCCAACGUCGCCUCUUGGCGUGCCAAAGUGGCAUUUGAUUUA .........-....(----(((((((((..((((.....--((....))..((((........))))..))).....(((....))))..)))))))))).......... ( -32.80) >DroSec_CAF1 14549 98 + 1 UACACAAUG-CUCAG----CCACUUUGGUGCUGGUACUC--GC----GCACUCGUAAAUCCCCGGGAAUCCAACGUCGCCUC-UGGCGUGCCAAAGUGGCAUUUGAUUUA .........-....(----(((((((((..((((..(((--(.----...............))))...))).....(((..-.))))..)))))))))).......... ( -30.89) >DroSim_CAF1 15813 80 + 1 UACACAAUG-CUCAG----CCACUUUGGUGCUGGUACUC--G------CACUCGUAAAU-----------------CGCCUCUUGGCGUGCCAAAGUGGCAUUUGAUUUA .........-....(----(((((((((..(.(((...(--(------....))...))-----------------)(((....))))..)))))))))).......... ( -29.00) >DroEre_CAF1 14523 96 + 1 UACACAAUG-CUCAG----CCACUUUGGUGCUGGUGCUC--G------CACUCGUAAAUCGCCG-GAAUCCAGUGUCGCCUCUUGGCGUGCCAAAGUGGCAUUUGAUUUA .........-....(----(((((((((((((((..((.--(------(...........)).)-)...)))))..((((....)))).))))))))))).......... ( -35.90) >DroYak_CAF1 15043 96 + 1 UACAAAAUG-CUCAG----CCACUUUGGUGCUGGUACUC--G------CACUCGUAAAUCGCCG-GAAUCCAGCGUCGCCUCUUGGCGUGCCAAAGUGGCAUUUGAUUUA .........-....(----(((((((((((((((..((.--(------(...........)).)-)...)))))..((((....)))).))))))))))).......... ( -37.00) >DroAna_CAF1 12777 100 + 1 UACAAGAGGACUCAGUCCACCACUUUGAUACUGGCACUCCAGCACUCGUACUCGUAAAUCCCCCGGAAUCUGACGUCGC----------GCCAAAGUGGCAUUUGAUUUA ..((((.((((...)))).((((((((...((((....))))....(((...(((...(((...))).....)))..))----------).))))))))..))))..... ( -24.00) >consensus UACACAAUG_CUCAG____CCACUUUGGUGCUGGUACUC__G______CACUCGUAAAUCCCCG_GAAUCCAACGUCGCCUCUUGGCGUGCCAAAGUGGCAUUUGAUUUA ...........((((....((((((((((((...........................((.....))..........(((....)))))))))))))))...)))).... (-19.57 = -20.52 + 0.94)



| Location | 19,571,884 – 19,571,987 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19571884 103 - 27905053 UAAAUCAAAUGCCACUUUGGCACGCCAAGAGGCGACGUUGGAUUCAUUGGGAUUUACGAGUGCGAGUGC--GAGUACCAGCACCAAAGUGG----CUAAG-CAUUGUGUA ..........((((((((((((((((....)))..(((.((((((....))))))))).))))..((((--........))))))))))))----)....-......... ( -36.70) >DroSec_CAF1 14549 98 - 1 UAAAUCAAAUGCCACUUUGGCACGCCA-GAGGCGACGUUGGAUUCCCGGGGAUUUACGAGUGC----GC--GAGUACCAGCACCAAAGUGG----CUGAG-CAUUGUGUA ....(((...((((((((((..((((.-..))))..(((((((((....))))).....((((----..--..)))))))).)))))))))----)))).-......... ( -35.50) >DroSim_CAF1 15813 80 - 1 UAAAUCAAAUGCCACUUUGGCACGCCAAGAGGCG-----------------AUUUACGAGUG------C--GAGUACCAGCACCAAAGUGG----CUGAG-CAUUGUGUA ....(((...(((((((((...((((....))))-----------------........(((------(--........))))))))))))----)))).-......... ( -27.30) >DroEre_CAF1 14523 96 - 1 UAAAUCAAAUGCCACUUUGGCACGCCAAGAGGCGACACUGGAUUC-CGGCGAUUUACGAGUG------C--GAGCACCAGCACCAAAGUGG----CUGAG-CAUUGUGUA ....(((...((((((((((((((((....)))..(.((((...)-))).)........)))------)--..((....))..))))))))----)))).-......... ( -31.00) >DroYak_CAF1 15043 96 - 1 UAAAUCAAAUGCCACUUUGGCACGCCAAGAGGCGACGCUGGAUUC-CGGCGAUUUACGAGUG------C--GAGUACCAGCACCAAAGUGG----CUGAG-CAUUUUGUA ....(((...((((((((((..((((....))))..(((((((((-((.((.....))..))------.--)))).))))).)))))))))----)))).-......... ( -37.70) >DroAna_CAF1 12777 100 - 1 UAAAUCAAAUGCCACUUUGGC----------GCGACGUCAGAUUCCGGGGGAUUUACGAGUACGAGUGCUGGAGUGCCAGUAUCAAAGUGGUGGACUGAGUCCUCUUGUA ..........((((((((((.----------.(..(((.((((((....))))))))).)..)..(((((((....))))))))))))))))((((...))))....... ( -32.30) >consensus UAAAUCAAAUGCCACUUUGGCACGCCAAGAGGCGACGUUGGAUUC_CGGGGAUUUACGAGUG______C__GAGUACCAGCACCAAAGUGG____CUGAG_CAUUGUGUA ....(((....(((((((((..((((....)))).......................(.(((............))))....))))))))).....)))........... (-16.68 = -16.83 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:30 2006