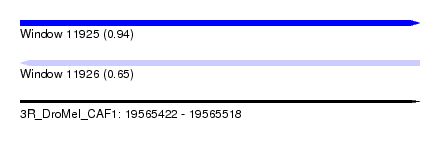
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,565,422 – 19,565,518 |
| Length | 96 |
| Max. P | 0.944491 |

| Location | 19,565,422 – 19,565,518 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565422 96 + 27905053 AUUCAAAUUUGCAACAAGCUGCCAGAAAU-CAGAGGCAGCCAACCAAAGUUGCCCACUUUACGAGCGCUUAUGUUGCAGAUGACAUGCGCCAACAAC ......((((((((((.((((((......-....))))))......((((.((.(.......).)))))).))))))))))................ ( -24.90) >DroSec_CAF1 8166 96 + 1 AUUCAAAUUUGCAACAAGCUGCCAGAAAU-CAGAGGCAGCCAACCAAAGUUGCCCACUUUACGAGCGCUUAUGUUGCAGAUGACAGGCGCCAACAAC ......((((((((((.((((((......-....))))))......((((.((.(.......).)))))).))))))))))................ ( -24.90) >DroSim_CAF1 9136 96 + 1 AUUCAGAUUUGCAACAAGCUGCCAGAAAU-CAGAGGCAGCCAACCAAAGUUGCCCACUUUACGAGCGCUUAUGUUGCAGAUGACAGGCGCCAACAAC ......((((((((((.((((((......-....))))))......((((.((.(.......).)))))).))))))))))................ ( -25.00) >DroEre_CAF1 8128 88 + 1 AUUCAAAUUUGCAACAAGCUGCCAGAAAG-CAGAGGCAGCCAACCAAAGUUGCCCACUUUACGAGCGCUUAUGUUGCAGAUGACAAGCG-------- ......(((((((((((((.((..(((((-..(.((((((........))))))).)))).)..)))))..))))))))))........-------- ( -27.00) >DroYak_CAF1 8210 88 + 1 AUUCAAAUUUGCAACAAGCUGCCAGAAAG-CAGAGGCAGCCAACCCAAGUUGCCCACUUUACGAGCGCUUAUGUGGCAGAUGACAGGCA-------- .........(((......((((((.((((-(.(.((((((........))))))).(((...))).)))).).)))))).......)))-------- ( -25.22) >DroAna_CAF1 6903 87 + 1 AUUCAAAUUUGCAACAAGCUGCCAGGAGUGGCGAGGCAGCCAACCA-GGUUGGUCGCUUUACUGCCGCUUAUGUUGCAGAUGACAGGC--------- ......((((((((((.........((((((((((((.((((((..-.)))))).)))))...))))))).)))))))))).......--------- ( -32.40) >consensus AUUCAAAUUUGCAACAAGCUGCCAGAAAU_CAGAGGCAGCCAACCAAAGUUGCCCACUUUACGAGCGCUUAUGUUGCAGAUGACAGGCG________ ......((((((((((.((((((...........))))))((((....))))...................))))))))))................ (-23.62 = -23.78 + 0.17)


| Location | 19,565,422 – 19,565,518 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565422 96 - 27905053 GUUGUUGGCGCAUGUCAUCUGCAACAUAAGCGCUCGUAAAGUGGGCAACUUUGGUUGGCUGCCUCUG-AUUUCUGGCAGCUUGUUGCAAAUUUGAAU ..((..(((((((((........))))..)))))..))((((..(((((.......(((((((....-......))))))).)))))..)))).... ( -27.80) >DroSec_CAF1 8166 96 - 1 GUUGUUGGCGCCUGUCAUCUGCAACAUAAGCGCUCGUAAAGUGGGCAACUUUGGUUGGCUGCCUCUG-AUUUCUGGCAGCUUGUUGCAAAUUUGAAU .....((((....))))..(((((((.....(((((.....)))))..........(((((((....-......))))))))))))))......... ( -27.70) >DroSim_CAF1 9136 96 - 1 GUUGUUGGCGCCUGUCAUCUGCAACAUAAGCGCUCGUAAAGUGGGCAACUUUGGUUGGCUGCCUCUG-AUUUCUGGCAGCUUGUUGCAAAUCUGAAU .....((((....))))..(((((((.....(((((.....)))))..........(((((((....-......))))))))))))))......... ( -27.70) >DroEre_CAF1 8128 88 - 1 --------CGCUUGUCAUCUGCAACAUAAGCGCUCGUAAAGUGGGCAACUUUGGUUGGCUGCCUCUG-CUUUCUGGCAGCUUGUUGCAAAUUUGAAU --------......(((..(((((((..((((((.(.((((((((((.((......)).)))))..)-))))).))).))))))))))....))).. ( -27.30) >DroYak_CAF1 8210 88 - 1 --------UGCCUGUCAUCUGCCACAUAAGCGCUCGUAAAGUGGGCAACUUGGGUUGGCUGCCUCUG-CUUUCUGGCAGCUUGUUGCAAAUUUGAAU --------(((....((.((((((...(((((((((.....))))).....((((.....))))..)-)))..))))))..))..)))......... ( -25.10) >DroAna_CAF1 6903 87 - 1 ---------GCCUGUCAUCUGCAACAUAAGCGGCAGUAAAGCGACCAACC-UGGUUGGCUGCCUCGCCACUCCUGGCAGCUUGUUGCAAAUUUGAAU ---------.....(((..(((((((..(((((((((....(((((....-.)))))))))))..((((....)))).))))))))))....))).. ( -33.70) >consensus ________CGCCUGUCAUCUGCAACAUAAGCGCUCGUAAAGUGGGCAACUUUGGUUGGCUGCCUCUG_AUUUCUGGCAGCUUGUUGCAAAUUUGAAU ..............(((..(((((((.....(((((.....)))))..........(((((((...........))))))))))))))....))).. (-25.35 = -25.43 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:25 2006