
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,565,081 – 19,565,362 |
| Length | 281 |
| Max. P | 0.995570 |

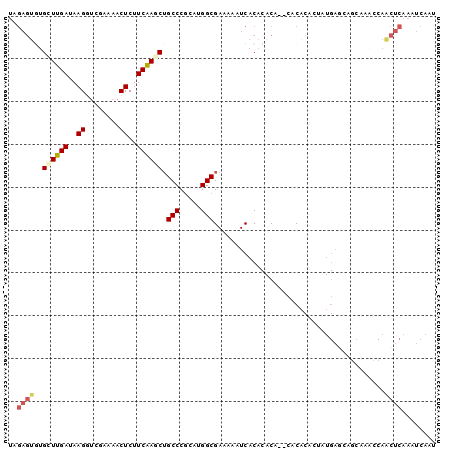
| Location | 19,565,081 – 19,565,174 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.46 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565081 93 + 27905053 ------G-GUUUGAUAAGGUCGAGAACUCUUCAAGCUGCCCGCAUGGCGAAAAAUCACACACACACACACACUAUGAGCAGGGAGCCAACUCAAAUCAAU ------(-((((((.......(((.....)))..(((.(((.(((((.(....................).)))))....))))))....)))))))... ( -17.55) >DroSec_CAF1 7832 97 + 1 UAGAGUGUGCUUGAUAAGGUCGGA-ACUCUUCAAACUGCCCGCAUGGCGAAAAAUCACACACA--CACACACUAUGAGCGGCAAACCAACUCAAAUCAAU ..(((((((..((((..(((..((-.....))..)))(((.....))).....))))..))))--)........((((.((....))..))))..))... ( -19.30) >DroSim_CAF1 8801 98 + 1 UAGAGUGUGCUUGAUAAGGUCGGAAACUCUUCAAACUGCCCGCAUGGCGAAAAAUCACACACA--CACACACUAUGAACAGCAAACCAACUCAAAUCAAU ....(((((..((((..(((..(((....)))..)))(((.....))).....))))..))))--).................................. ( -16.40) >DroEre_CAF1 7788 83 + 1 UGGAGUAUGCUUGAUAAGCUCGAAAACUCUUCGAGCGGCCCACAUGGCGAAAAAUCACA--CA--CUCGCCAUG-------------AGCUCAAAUCAAU ..((((...........(((((((.....)))))))......((((((((.........--..--.))))))))-------------.))))........ ( -27.20) >DroYak_CAF1 7843 92 + 1 UAGAGUAUGCUUGAUAAG------AACUCUUCGAGCUGCCCGCAUGGCGAAAAAUCACACACA--CUCACCAUGUGAGCACCGAAGCAACUCAAAUCAAU ..((((..((((((..((------....))))))))(((.(((((((.((.............--.)).))))))).)))........))))........ ( -25.34) >consensus UAGAGUGUGCUUGAUAAGGUCGAAAACUCUUCAAGCUGCCCGCAUGGCGAAAAAUCACACACA__CACACACUAUGAGCAGCAAACCAACUCAAAUCAAU ..((((..((((((..((........))..)))))).(((.....)))........................................))))........ (-11.70 = -12.46 + 0.76)


| Location | 19,565,081 – 19,565,174 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -11.72 |
| Energy contribution | -13.22 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565081 93 - 27905053 AUUGAUUUGAGUUGGCUCCCUGCUCAUAGUGUGUGUGUGUGUGUGAUUUUUCGCCAUGCGGGCAGCUUGAAGAGUUCUCGACCUUAUCAAAC-C------ .(((((..(.(((((......((((..(((.(((.((((((.((((....)))))))))).))))))....))))..))))))..)))))..-.------ ( -26.50) >DroSec_CAF1 7832 97 - 1 AUUGAUUUGAGUUGGUUUGCCGCUCAUAGUGUGUG--UGUGUGUGAUUUUUCGCCAUGCGGGCAGUUUGAAGAGU-UCCGACCUUAUCAAGCACACUCUA .(((((..(.(((((...(((..(((.(.(((.((--((((.((((....)))))))))).))).).))).).))-.))))))..))))).......... ( -29.90) >DroSim_CAF1 8801 98 - 1 AUUGAUUUGAGUUGGUUUGCUGUUCAUAGUGUGUG--UGUGUGUGAUUUUUCGCCAUGCGGGCAGUUUGAAGAGUUUCCGACCUUAUCAAGCACACUCUA .(((((..(.(((((...(((.((((.(.(((.((--((((.((((....)))))))))).))).).)))).)))..))))))..))))).......... ( -35.60) >DroEre_CAF1 7788 83 - 1 AUUGAUUUGAGCU-------------CAUGGCGAG--UG--UGUGAUUUUUCGCCAUGUGGGCCGCUCGAAGAGUUUUCGAGCUUAUCAAGCAUACUCCA ..((.((((((((-------------(((((((((--..--........)))))))))..))).((((((((...))))))))...)))))))....... ( -27.80) >DroYak_CAF1 7843 92 - 1 AUUGAUUUGAGUUGCUUCGGUGCUCACAUGGUGAG--UGUGUGUGAUUUUUCGCCAUGCGGGCAGCUCGAAGAGUU------CUUAUCAAGCAUACUCUA .(((((..(((...(((((((((((.(((((((((--............))))))))).))))).).)))))..))------)..))))).......... ( -33.00) >consensus AUUGAUUUGAGUUGGUUCGCUGCUCAUAGUGUGUG__UGUGUGUGAUUUUUCGCCAUGCGGGCAGCUUGAAGAGUUUCCGACCUUAUCAAGCACACUCUA .......(((((.((....)))))))...(((.((...(((.((((....))))))).)).)))((((((.(((........))).))))))........ (-11.72 = -13.22 + 1.50)

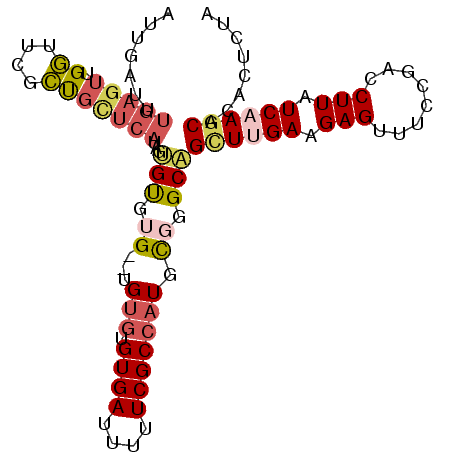
| Location | 19,565,105 – 19,565,198 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -16.36 |
| Consensus MFE | -10.37 |
| Energy contribution | -9.99 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565105 93 + 27905053 CAAGCUGCCCGCAUGGCGAAAAAUCACACACACACACACACUAUGAGCAGGGAGCCAACUCAAAUCAAUUUCUGUUUAAGCCAGA---------------AAAAGGCA ......(((....((((((....))..................(((((((..(...............)..))))))).))))..---------------....))). ( -18.36) >DroSec_CAF1 7862 91 + 1 CAAACUGCCCGCAUGGCGAAAAAUCACACACA--CACACACUAUGAGCGGCAAACCAACUCAAAUCAAUUUCUGUUUAAGCCAGA---------------AAAAGGCA ......(((....((((((....)).......--.........((((.((....))..)))).................))))..---------------....))). ( -14.20) >DroSim_CAF1 8832 91 + 1 CAAACUGCCCGCAUGGCGAAAAAUCACACACA--CACACACUAUGAACAGCAAACCAACUCAAAUCAAUUUCUGUUUAAGCCAGA---------------AAAAGGCA ......(((....((((((....)).......--.........(((((((.(((..............)))))))))).))))..---------------....))). ( -14.34) >DroYak_CAF1 7868 106 + 1 CGAGCUGCCCGCAUGGCGAAAAAUCACACACA--CUCACCAUGUGAGCACCGAAGCAACUCAAAUCAAUUUCUGUUUAAGCCACAAAAAAAAAAAAUGAAAAAAGGCU ((.(.(((.(((((((.((.............--.)).))))))).))))))(((((...............))))).((((......................)))) ( -18.55) >consensus CAAACUGCCCGCAUGGCGAAAAAUCACACACA__CACACACUAUGAGCAGCAAACCAACUCAAAUCAAUUUCUGUUUAAGCCAGA_______________AAAAGGCA ......(((....((((((....))..................(((((((.((...............)).))))))).)))).....................))). (-10.37 = -9.99 + -0.37)

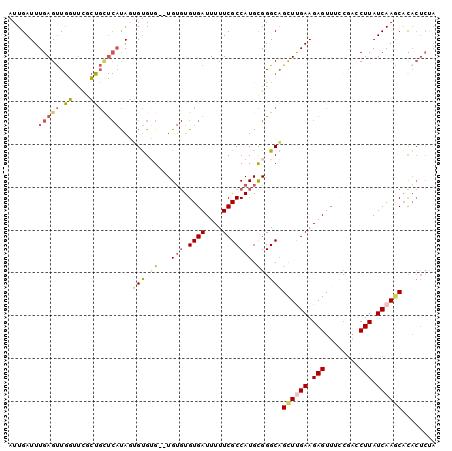
| Location | 19,565,105 – 19,565,198 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565105 93 - 27905053 UGCCUUUU---------------UCUGGCUUAAACAGAAAUUGAUUUGAGUUGGCUCCCUGCUCAUAGUGUGUGUGUGUGUGUGAUUUUUCGCCAUGCGGGCAGCUUG ...(..((---------------((((.......))))))..)...(((((.((...)).))))).(((.(((.((((((.((((....)))))))))).)))))).. ( -26.70) >DroSec_CAF1 7862 91 - 1 UGCCUUUU---------------UCUGGCUUAAACAGAAAUUGAUUUGAGUUGGUUUGCCGCUCAUAGUGUGUG--UGUGUGUGAUUUUUCGCCAUGCGGGCAGUUUG ...(..((---------------((((.......))))))..)...(((((.((....)))))))...(((.((--((((.((((....)))))))))).)))..... ( -27.00) >DroSim_CAF1 8832 91 - 1 UGCCUUUU---------------UCUGGCUUAAACAGAAAUUGAUUUGAGUUGGUUUGCUGUUCAUAGUGUGUG--UGUGUGUGAUUUUUCGCCAUGCGGGCAGUUUG .(((....---------------...)))...(((((((((..((....))..)))).)))))((.(.(((.((--((((.((((....)))))))))).))).).)) ( -26.80) >DroYak_CAF1 7868 106 - 1 AGCCUUUUUUCAUUUUUUUUUUUUGUGGCUUAAACAGAAAUUGAUUUGAGUUGCUUCGGUGCUCACAUGGUGAG--UGUGUGUGAUUUUUCGCCAUGCGGGCAGCUCG ........................(..((((((((((...))).)))))))..)...(.(((((.(((((((((--............))))))))).))))).)... ( -30.60) >consensus UGCCUUUU_______________UCUGGCUUAAACAGAAAUUGAUUUGAGUUGGUUCGCUGCUCAUAGUGUGUG__UGUGUGUGAUUUUUCGCCAUGCGGGCAGCUUG .((((...................(..((((((((((...))).)))))))..).......................((((((((....))).)))))))))...... (-19.60 = -19.22 + -0.38)


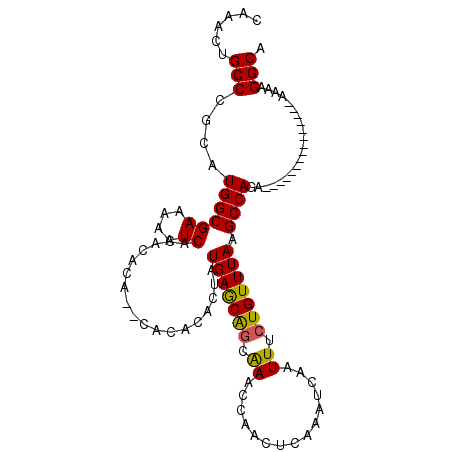
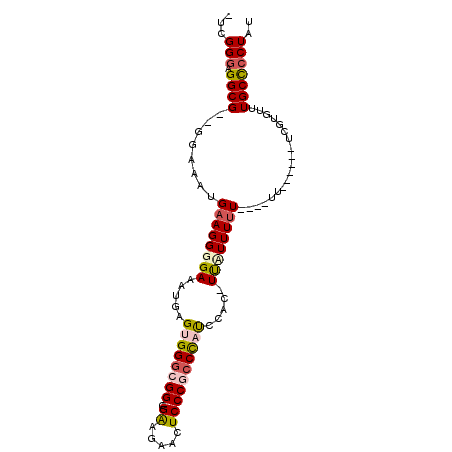
| Location | 19,565,198 – 19,565,292 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565198 94 + 27905053 GACGGGUGGCG--GGAAAUGAAGGGGAAAUGAGUGGGCGGAAGAAGAACUCCCGACCUUCCAC-UUUUUUUUUUUUUUCGU---UCGUGUUUGCCCCUAU ...(((..(((--.(((.(((((((((((.((((((((((.((.....)).)))....)))))-))...))))))))))))---)).)).)..))).... ( -31.40) >DroSec_CAF1 7953 83 + 1 -UCGGGAGGCG--GGAAAUGAAGGGGAAAUGAGUGGGCGG-UGAAGAACUCCCGCCCACCCAC-UUCUUUUU------------UCGUGUUUGCCCCUAU -..(((.((((--((....(((((((((.((.((((((((-.((.....)))))))))).)).-))))))))------------)....))))))))).. ( -38.00) >DroSim_CAF1 8923 83 + 1 -UCGGGAGGCG--GGAAAUGAAGGGGAAAUGAGUGGGCGG-CGAAGAACUCCCGCCCAUCUAC-UUCUUUUU------------UCGUGUUUGCCCCUAU -..(((.((((--((....(((((((((.((..(((((((-.((.....)))))))))..)).-))))))))------------)....))))))))).. ( -34.80) >DroEre_CAF1 7911 85 + 1 -UCGGGAGGCGCGGAAAUUGAAGGGGAAAUGAGUGGGCGG-CGGCGAACUCCCUCCU-CAGUUUUCAUUUUU----UU--------UUUUCUGCUGCUAU -......((((((((((..((((((((((((((.(((.((-.(.....).)))))))-))..)))).)))))----).--------.))))))).))).. ( -26.70) >DroYak_CAF1 7974 94 + 1 -UUGGGAGGCGCGGAAAUUGCAGGGGAAAUGAGUGGGCGG-UGGAGAACUCCCUCCCAUCGUUUUCAUUUCU----UUCGUUUUUGGUUUUUGCCCCUAU -.((((.((((..((....((..((((((((((..(((((-(((.((......)))))))))))))))))))----)..))......))..)))))))). ( -34.60) >consensus _UCGGGAGGCG__GGAAAUGAAGGGGAAAUGAGUGGGCGG_CGAAGAACUCCCGCCCAUCCAC_UUAUUUUU____UU______UCGUGUUUGCCCCUAU ...(((.((((........((((((((.....((((((((..((.....)))))))))).....))))))))...................))))))).. (-15.57 = -16.25 + 0.68)


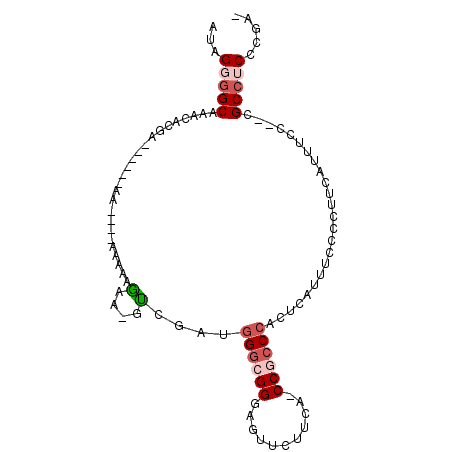
| Location | 19,565,198 – 19,565,292 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -8.80 |
| Energy contribution | -9.64 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565198 94 - 27905053 AUAGGGGCAAACACGA---ACGAAAAAAAAAAAAAA-GUGGAAGGUCGGGAGUUCUUCUUCCGCCCACUCAUUUCCCCUUCAUUUCC--CGCCACCCGUC ..(((((.((((((..---.................-)))...((.((((((.....)))))).)).....))))))))........--........... ( -20.31) >DroSec_CAF1 7953 83 - 1 AUAGGGGCAAACACGA------------AAAAAGAA-GUGGGUGGGCGGGAGUUCUUCA-CCGCCCACUCAUUUCCCCUUCAUUUCC--CGCCUCCCGA- ...(((((......((------------(....(((-(((((((((((((((...))).-)))))))))))))))...)))......--.)))))....- ( -35.22) >DroSim_CAF1 8923 83 - 1 AUAGGGGCAAACACGA------------AAAAAGAA-GUAGAUGGGCGGGAGUUCUUCG-CCGCCCACUCAUUUCCCCUUCAUUUCC--CGCCUCCCGA- ...(((((......((------------(....(((-((((.((((((((((...))).-))))))))).)))))...)))......--.)))))....- ( -25.32) >DroEre_CAF1 7911 85 - 1 AUAGCAGCAGAAAA--------AA----AAAAAUGAAAACUG-AGGAGGGAGUUCGCCG-CCGCCCACUCAUUUCCCCUUCAAUUUCCGCGCCUCCCGA- ...((.((.((((.--------..----.....(....).((-(((.(((((.......-............)))))))))).)))).)))).......- ( -14.41) >DroYak_CAF1 7974 94 - 1 AUAGGGGCAAAAACCAAAAACGAA----AGAAAUGAAAACGAUGGGAGGGAGUUCUCCA-CCGCCCACUCAUUUCCCCUGCAAUUUCCGCGCCUCCCAA- ...(((((................----.(((((((....(.((((..(((....))).-...))))))))))))....((.......)))))))....- ( -23.70) >consensus AUAGGGGCAAACACGA______AA____AAAAAGAA_GUCGAUGGGCGGGAGUUCUUCA_CCGCCCACUCAUUUCCCCUUCAUUUCC__CGCCUCCCGA_ ...(((((.........................(....)....((((((...........))))))........................)))))..... ( -8.80 = -9.64 + 0.84)


| Location | 19,565,256 – 19,565,362 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19565256 106 + 27905053 CAC-UUUUUUUUUUUUUUCGU---UCGUGUUUGCCCCUAUCUCAGCAUGUUAUCCAUGAAGCCGUGUAAUAUGACUGUGAUUGGAAAUUGAAAUUGCAUCCGGCAAACAU ...-.................---..(((((((((((.(((.((((((((((..((((....))))))))))).))).))).))....((......))...))))))))) ( -27.40) >DroSec_CAF1 8009 97 + 1 CAC-UUCUUUUU------------UCGUGUUUGCCCCUAUCUCAGCAUGUAAUCCAUGAAGCCGUGUAAUAUGACUGUGAUUGGAAAUUGAAAUUGCAUCCGGCAAACAU ...-........------------..(((((((((((.(((.((((((((....((((....))))..))))).))).))).))....((......))...))))))))) ( -24.80) >DroSim_CAF1 8979 97 + 1 UAC-UUCUUUUU------------UCGUGUUUGCCCCUAUCUCAGCAUGUAAUCCAUGAAGCCGUGUAAUAUGACUGUGAUUGGAAAUUGAAAUUGCAUCCGGCAAACAU ...-........------------..(((((((((((.(((.((((((((....((((....))))..))))).))).))).))....((......))...))))))))) ( -24.80) >DroEre_CAF1 7968 98 + 1 GUUUUCAUUUUU----UU--------UUUUCUGCUGCUAUCUCAGCGUGUAAUCCAUGAAGCCGUGUAAUAUGACUGCGAUUGGAAAUUGAAAUUGCAUCCGGCAAACAU ((((........----..--------......((((......))))..............((((.((......))(((((((.........)))))))..)))))))).. ( -16.70) >DroYak_CAF1 8032 106 + 1 GUUUUCAUUUCU----UUCGUUUUUGGUUUUUGCCCCUAUCUCGGCAUGUAAUCCAUGAAGCCGUGUAAUAUGACUGUGAUUGGAAAUUGAAAUUGCAUCCGGCAAACAU ..(((((.....----.........(((....)))((.(((.((((((((....((((....))))..))))).))).))).))....)))))((((.....)))).... ( -20.10) >DroAna_CAF1 6753 80 + 1 C------------------------------UGCCAGCCAGCCAGCUUGUAAUCCAUGAAGCCGUGUAAUAUGACUGUGAUUGGAAAUUGAAAUUGCAUCCGGCAGACAU (------------------------------((((..((((((((.(..((...((((....))))...))..)))).).))))....((......))...))))).... ( -17.70) >consensus CAC_UUAUUUUU____________UCGUGUUUGCCCCUAUCUCAGCAUGUAAUCCAUGAAGCCGUGUAAUAUGACUGUGAUUGGAAAUUGAAAUUGCAUCCGGCAAACAU .............................((((((...(((.((((((((....((((....))))..))))).))).))).(((...((......)))))))))))... (-15.32 = -16.10 + 0.78)

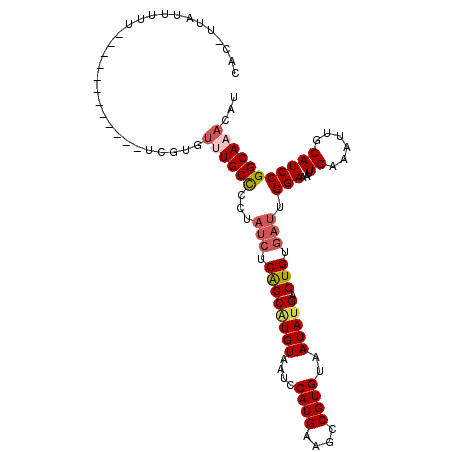
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:23 2006