
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,542,043 – 19,542,141 |
| Length | 98 |
| Max. P | 0.547683 |

| Location | 19,542,043 – 19,542,141 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
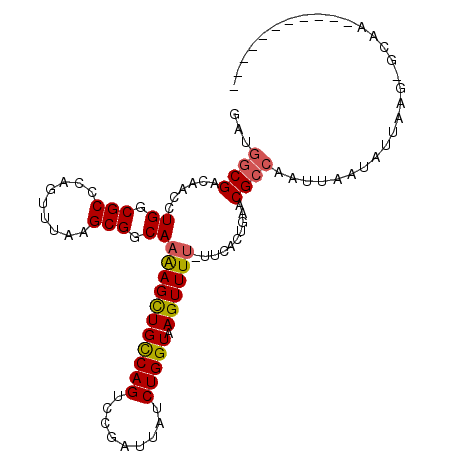
>3R_DroMel_CAF1 19542043 98 - 27905053 GAUGGCGACAACCUGGCGCCCAGUUUGAGCGGCAAAAGCUGCCAGUCCGAUUAUCUGGUAAGUUUUGUCCACUAACCGCCAAUUAAUAUUAAGCCCAC----------- ..(((((.(((.((((...)))).)))((.(((((((..((((((.........))))))..))))).)).))...))))).................----------- ( -28.90) >DroVir_CAF1 10391 99 - 1 GAUGGCGACAAUCUGGCGCCCAGUUUAAGCGGCAAGAGCUGCCAGUCCGAUUAUCUGGUAAGUUUU---UGCUGAACGCCAAUUAAUAUUAAAGGCAAUGGC------- ..(((((.....((((...))))......((((((((((((((((.........))))).))))))---)))))..))))).....................------- ( -31.90) >DroGri_CAF1 11497 90 - 1 GAUGGCGACAAUCUGGCGCCCAGUUUGAGCGGCAAGAGUUGUCAGUCUGAUUAUCUGGUAAGUUUU-GUUGCUGAACGCCAAUUAAUAAUA------------------ ..(((((.(((.((((...)))).)))((((((((((.((..(((.........)))..)).))))-))))))...)))))..........------------------ ( -32.40) >DroMoj_CAF1 12617 100 - 1 GACGGCGACAAUCUGGCGCCCAGUUUAAGCGGCAAGAGCUGCCAGUCCGAUUAUCUGGUAAGUUUU-G-CGCUGAACGCCAAUUAAUAUUAAAGGCAACGGC------- ...((((.(.....).))))..(((((.(((.(((((..((((((.........))))))..))))-)-))))))))(((.............)))......------- ( -29.52) >DroAna_CAF1 10782 93 - 1 GACACCGACAACCUGGCGCCGAGUUUAAGCGGCAAAAGCUGCCAGUCCGAUUAUCUGGUAAGUUU--UUCACUGAACGCCAAUUAAUAUUUAG---AG----------- .............((((((((........))))((((((((((((.........))))).)))))--))........))))............---..----------- ( -23.50) >DroPer_CAF1 9630 107 - 1 GAUGGCGACAACCUGGCGCCGAGUCUAAGCGGCAAAAGCUGCCAGUCCGAUUAUCUGGUAAGUUUU-UUCACUGAACGCCAAUUAAUAUUAAGAGUAA-AGUAAAGGAG ..(((((..........((((........))))((((((((((((.........))))).))))))-)........))))).................-.......... ( -25.70) >consensus GAUGGCGACAACCUGGCGCCCAGUUUAAGCGGCAAAAGCUGCCAGUCCGAUUAUCUGGUAAGUUUU_UUCACUGAACGCCAAUUAAUAUUAAG_GCAA___________ ...((((......((.(((.........))).))(((((((((((.........))))).))))))..........))))............................. (-18.61 = -18.58 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:12 2006