| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,523,061 – 19,523,221 |
| Length | 160 |
| Max. P | 0.683578 |

| Location | 19,523,061 – 19,523,181 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19523061 120 + 27905053 AAUGAAAACGGUGCCCAGCUGCCACUUGUUAGGCUUUAGAUACUGGAUUAGCCAGAUGGGCAGACAGGGCUCCAGCAUGGCCAUUGUUGAGGUUGAAAACCAAAUGGCCAGCACAAUGGC .........((.((((..(((((....(((.((((..............)))).))).)))))...)))).)).((.((((((((.....(((.....))).))))))))))........ ( -43.14) >DroVir_CAF1 15813 120 + 1 UAUGAAAACUGUGCCCAAUUGCCAUUUAUUUGGAUGCAGAUAUUGAAUCAACCAAAUGGGCAAACACGGCUCCAGUAUGGCCAUUGUGGAAGUGGAUAGCCAAAUGGCGAUCACAAUGGC .......((((.(((...(((((...(((((((.((............)).))))))))))))....)))..))))...((((((((((.........(((....)))..)))))))))) ( -35.00) >DroGri_CAF1 16689 120 + 1 AAUAAAAACCGUGCCCAAUUGCCAUUUAUUUGGGUGCAAGUACUGUAUCAACCAGAUGGGCAAACACGGCUCCAAUAUGGCCAUUGUGGAUGUGGACAGCCAAAUGGCAACCACAAUGGA ........(((((.....(((((...((((((((((((.....)))))...)))))))))))).)))))...........(((((((((.((....))(((....)))..))))))))). ( -39.00) >DroWil_CAF1 14387 120 + 1 AAUGAAAACUGUUCCCAGCUGCCACUUGUUCGGCUUGAGAUAUUGAAUUAACCAAAUGGGCAGACACGGUUCCAAUAUGGCCAUUGUUGAUGUGGAGAACCAAAUCGCCACAACAAUGGC ......((((((......(((((....(((.((.((((.........)))))).))).)))))..))))))........((((((((...(((((.((......)).))))))))))))) ( -31.60) >DroMoj_CAF1 15096 120 + 1 UAUGAAAACGGUGCCCAAUUGCCACUUGUUCGGAUGGAGAUACUGAAUCAGCCAGAUGGGCAAACAGGGCUCCAAUAUGGCCAUUGUGGAUGUGGACAACCAAAUGGCGAUGACAAUGGC .........((.((((..(((((.((.((((((((....)).)))))).)).......)))))...)))).))..(((.((((((.(((.((....)).))))))))).)))........ ( -35.80) >DroAna_CAF1 16278 120 + 1 UAUGAAGACCGUUCCCAACUGCCACUUGUUAGGCUUCAGAUACUGAAUUAGCCAAAUUGGCAGACAAGGCUCUAGCAUGGCCAUGGUGGAGGUGGAGAACCAAAUGGCCAGCACAAUGGC ..........(((((((.((((((...(((.((((((((...))))...)))).)))))))))......(((((.(((....))).))))).))).))))......((((......)))) ( -37.90) >consensus AAUGAAAACCGUGCCCAACUGCCACUUGUUAGGCUGCAGAUACUGAAUCAACCAAAUGGGCAAACACGGCUCCAAUAUGGCCAUUGUGGAUGUGGACAACCAAAUGGCCACCACAAUGGC ..........(.(((...(((((....(((.((..................)).))).)))))....))).).......((((((((((.(((.............))).)))))))))) (-23.00 = -23.17 + 0.17)


| Location | 19,523,101 – 19,523,221 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19523101 120 + 27905053 UACUGGAUUAGCCAGAUGGGCAGACAGGGCUCCAGCAUGGCCAUUGUUGAGGUUGAAAACCAAAUGGCCAGCACAAUGGCCAGGAUCAUUUUGCAUUCGAGCAGCGAUCGCCACUUGGGU ..(((((...(((...((......)).))))))))..((((((((((...((((...........))))...)))))))))).((((...((((......)))).))))(((.....))) ( -38.30) >DroVir_CAF1 15853 120 + 1 UAUUGAAUCAACCAAAUGGGCAAACACGGCUCCAGUAUGGCCAUUGUGGAAGUGGAUAGCCAAAUGGCGAUCACAAUGGCCAAAAUCAUUUUGCAUUCAAGAAGCGCACGCCACUUAGUC ..(((((((((.....(((((.......)).)))...((((((((((((.........(((....)))..))))))))))))........))).))))))...((....))......... ( -32.80) >DroGri_CAF1 16729 120 + 1 UACUGUAUCAACCAGAUGGGCAAACACGGCUCCAAUAUGGCCAUUGUGGAUGUGGACAGCCAAAUGGCAACCACAAUGGACAGUAUCAUUUUGGAUUCAAGGAGCGUGCCCCAUUUGGUC ..........((((((((((....((((.((((....((.(((((((((.((....))(((....)))..))))))))).))(.(((......))).)..)))))))).)))))))))). ( -47.80) >DroWil_CAF1 14427 120 + 1 UAUUGAAUUAACCAAAUGGGCAGACACGGUUCCAAUAUGGCCAUUGUUGAUGUGGAGAACCAAAUCGCCACAACAAUGGCCAGAAUCAUUUUACAUUCUAGCAACGGGCGCCAUUUGGGU ...........((((((((((......(((((.....((((((((((...(((((.((......)).))))))))))))))))))))..........((......)))).)))))))).. ( -41.00) >DroMoj_CAF1 15136 120 + 1 UACUGAAUCAGCCAGAUGGGCAAACAGGGCUCCAAUAUGGCCAUUGUGGAUGUGGACAACCAAAUGGCGAUGACAAUGGCCAAAAUCAUUUUGCAUUCGAGCAACGCGCGCCAUUUGGUG ..........(((((((((((......((((....(((.((((((.(((.((....)).))))))))).))).....)))).........((((......))))...)).))))))))). ( -39.70) >DroAna_CAF1 16318 120 + 1 UACUGAAUUAGCCAAAUUGGCAGACAAGGCUCUAGCAUGGCCAUGGUGGAGGUGGAGAACCAAAUGGCCAGCACAAUGGCCAGAAUCAUUUUACACUCAAGCAGGGGUCGCCACUUCGGG ..(((((...(((.....)))......((((((.((...((....)).(((((((((.......((((((......))))))......)))))).)))..)).)))))).....))))). ( -38.22) >consensus UACUGAAUCAACCAAAUGGGCAAACACGGCUCCAAUAUGGCCAUUGUGGAUGUGGACAACCAAAUGGCCACCACAAUGGCCAGAAUCAUUUUGCAUUCAAGCAGCGCGCGCCACUUGGGC ...........((((...(((......(....)....((((((((((((.(((.............))).))))))))))))...........................)))..)))).. (-22.85 = -23.77 + 0.92)

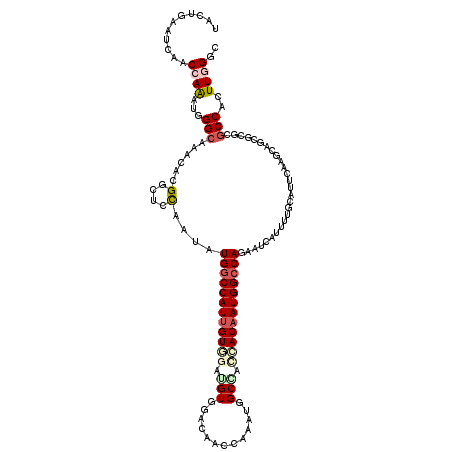

| Location | 19,523,101 – 19,523,221 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -23.06 |
| Energy contribution | -24.25 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19523101 120 - 27905053 ACCCAAGUGGCGAUCGCUGCUCGAAUGCAAAAUGAUCCUGGCCAUUGUGCUGGCCAUUUGGUUUUCAACCUCAACAAUGGCCAUGCUGGAGCCCUGUCUGCCCAUCUGGCUAAUCCAGUA .....(((((((((((.(((......)))...)))))...))))))....((((((((.(((.....))).....))))))))(((((((.....(((.........)))...))))))) ( -36.60) >DroVir_CAF1 15853 120 - 1 GACUAAGUGGCGUGCGCUUCUUGAAUGCAAAAUGAUUUUGGCCAUUGUGAUCGCCAUUUGGCUAUCCACUUCCACAAUGGCCAUACUGGAGCCGUGUUUGCCCAUUUGGUUGAUUCAAUA ((((((((((((.((((((((((....)))........(((((((((((((.(((....))).))).......))))))))))....)))).)))...)).))))))))))......... ( -38.81) >DroGri_CAF1 16729 120 - 1 GACCAAAUGGGGCACGCUCCUUGAAUCCAAAAUGAUACUGUCCAUUGUGGUUGCCAUUUGGCUGUCCACAUCCACAAUGGCCAUAUUGGAGCCGUGUUUGCCCAUCUGGUUGAUACAGUA (((((.(((((((((((((((((....)))...((((.((.(((((((((..(((....)))((....)).))))))))).)))))))))).)))))...))))).)))))......... ( -45.00) >DroWil_CAF1 14427 120 - 1 ACCCAAAUGGCGCCCGUUGCUAGAAUGUAAAAUGAUUCUGGCCAUUGUUGUGGCGAUUUGGUUCUCCACAUCAACAAUGGCCAUAUUGGAACCGUGUCUGCCCAUUUGGUUAAUUCAAUA ..((((((((.((...((((......)))).(((.((((((((((((((((((.((......)).)))))...))))))))).....)))).)))....))))))))))........... ( -40.30) >DroMoj_CAF1 15136 120 - 1 CACCAAAUGGCGCGCGUUGCUCGAAUGCAAAAUGAUUUUGGCCAUUGUCAUCGCCAUUUGGUUGUCCACAUCCACAAUGGCCAUAUUGGAGCCCUGUUUGCCCAUCUGGCUGAUUCAGUA (((((.((((.(((((..((((.((((...........((((((((((....(((....)))((....))...)))))))))))))).))))..)))..)))))).))).))........ ( -35.80) >DroAna_CAF1 16318 120 - 1 CCCGAAGUGGCGACCCCUGCUUGAGUGUAAAAUGAUUCUGGCCAUUGUGCUGGCCAUUUGGUUCUCCACCUCCACCAUGGCCAUGCUAGAGCCUUGUCUGCCAAUUUGGCUAAUUCAGUA .(((((.(((((((....((((((((........))))(((((((.(((..((.....(((....)))))..))).))))))).....))))...))).)))).)))))........... ( -37.10) >consensus AACCAAAUGGCGCCCGCUGCUUGAAUGCAAAAUGAUUCUGGCCAUUGUGAUGGCCAUUUGGUUCUCCACAUCCACAAUGGCCAUACUGGAGCCGUGUCUGCCCAUCUGGCUAAUUCAGUA ..(((.((((((..((..((((.((((...........(((((((((((...(((....)))..........))))))))))))))).))))..))..)))).)).)))........... (-23.06 = -24.25 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:07 2006