
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,522,765 – 19,522,901 |
| Length | 136 |
| Max. P | 0.861108 |

| Location | 19,522,765 – 19,522,884 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -38.21 |
| Consensus MFE | -24.81 |
| Energy contribution | -25.97 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19522765 119 + 27905053 GCAUCCACAAAGGUGGCCAAGAGUGGAACCAAUGCGGCAUCGAUUACUCCAAUACCCAGUCCCAAGGCAAAAUGGGGCAGCAAUAGUUGGGCCACCGUUGUAGCACUGGGUAUCUGCAA- (((.((.((..(((..(((....))).)))..)).))..............(((((((((((((........))))((.(((((....((....))))))).))))))))))).)))..- ( -36.00) >DroVir_CAF1 15524 120 + 1 GCAUCCACAAAAGUGGCCAGAAGUGGAACCAAGGCCGCAUCGAUCACACCAAUGCCUAGGCCCAGGGCAAAGUGGGGCAACAGUAGUUGAGCAACAGUCGUGGCACUGGGUAUCUGCAUA ....((((....))))......(..(((((.(((((((.(((((.((.(((.(((((.......)))))...)))(....).)).)))))((....)).))))).)).))).))..)... ( -37.70) >DroGri_CAF1 16404 120 + 1 GCAUCCACACAACUGGCCAACAGUGGCACCAAAGCUGCAUCAAUUACACCAAUGCCCAGGCCCAGGGCAAAAUGGGGCAACAGUAACUGAGCCACAGUUGUGGCGCUGGGAAUCUGCAUU ((((((.((((((((((((....)))).........((.(((.((((.(((.(((((.......)))))...)))(....).)))).)))))..))))))))......)))...)))... ( -41.10) >DroWil_CAF1 14099 119 + 1 GCAUCUACAAAGCUAGCCAGCAGUGGAACUAAAGCAGCAUCAAUUAUUCCUAUGCCCAAUCCCAAUGCAAAAUGGGGCAACAGAAGCUGUGCCAGUGUUGUGGCACUGGGUAUCUGCAA- ...(((((...(((....))).)))))......((((..............(((((((.(((((........)))))...........((((((......)))))))))))))))))..- ( -32.93) >DroMoj_CAF1 14815 119 + 1 GCAUCCACAAAGGUAGCCAGGAGUGGAACCAAGGCCGCAUCAAUCACCCCAAUGCCCAAGCCCAGAGCAAAAUGGGGCAGCAAGAGCUGCGCCACAGUUGUGGCGCUGGGUAUCUGCAA- (((........((.......((((((........)))).)).......)).(((((((.(((((.(((....(((.(((((....))))).)))..))).))).))))))))).)))..- ( -43.44) >DroAna_CAF1 15991 119 + 1 GCAUCCACAAAGGUGGCCAGCAGGGGAACCAGAGCAGCAUCAAUAACGCCAAUGCCCAGACCCAGAGCAAAGUGGGGUAACAGCAAUUGGGCCACUGUUGUGGCACUGGGAAUCUAUAA- ....((((((.(((((((.((.((....)).((......))......))((((((....((((...((...)).))))....)).))))))))))).))))))................- ( -38.10) >consensus GCAUCCACAAAGGUGGCCAGCAGUGGAACCAAAGCAGCAUCAAUUACACCAAUGCCCAGGCCCAGGGCAAAAUGGGGCAACAGUAGCUGAGCCACAGUUGUGGCACUGGGUAUCUGCAA_ ((((((((..............)))))........................((((((((.((((........))))..............(((((....))))).)))))))).)))... (-24.81 = -25.97 + 1.17)

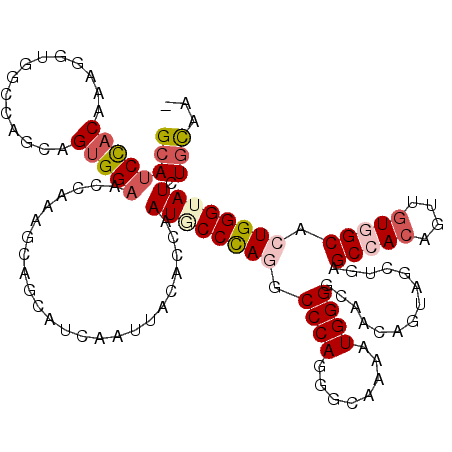
| Location | 19,522,805 – 19,522,901 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19522805 96 + 27905053 CGAUUACUCCAAUACCCAGUCCCAAGGCAAAAUGGGGCAGCAAUAGUUGGGCCACCGUUGUAGCACUGGGUAUCUGCAAAG---UAAAUAA-UAUAUUUG ...(((((.(((((((((((((((........))))((.(((((....((....))))))).))))))))))).))...))---)))....-........ ( -27.50) >DroSec_CAF1 17587 96 + 1 CGAUUACUCCAAUACCCAGUCCCAAGGCAAAAUGGGGCAGCAAUAGUUGGGCCACUGUUGUAGCACUGGGUAUCUGCAAAG---UAAAUAA-UAUAUAUG ...(((((.(((((((((((((((........))))((.((((((((......)))))))).))))))))))).))...))---)))....-........ ( -31.50) >DroSim_CAF1 17776 96 + 1 CGAUUACUCCAAUACCCAGUCCCAAGGCAAAAUGGGGCAGCAAUAGUUGGGCCACUGUUGUAGCACUGGGUAUCUGCAAAG---UAAAUAA-UAUAUAUG ...(((((.(((((((((((((((........))))((.((((((((......)))))))).))))))))))).))...))---)))....-........ ( -31.50) >DroEre_CAF1 17713 97 + 1 CGAUUACACCAAUGCCCAGGCCCAAGGCGAAAUGGGGCAUCAAUAGCUGGGCAACCGUCGUGGCACUGGGUAUCUGUAAAA---UUCAGAGUUAUAAUUG ((((((......((((((((((...(((.......(((.......)))((....)))))..))).)))))))((((.....---..))))....)))))) ( -31.60) >DroYak_CAF1 17797 97 + 1 CGAUUACACCAAUGCCCAGGCCCAAGGCGAAAUGGGGCAUCAGCAGUUGGGCAACCGUUGUAGCACUCGGUAUCUGUAAGG---UUCAGAGUUAUAAUUG ((((((......(((((((.((((........))))((....))..)))))))((((..(.....).)))).((((.....---..))))....)))))) ( -24.80) >DroMoj_CAF1 14855 98 + 1 CAAUCACCCCAAUGCCCAAGCCCAGAGCAAAAUGGGGCAGCAAGAGCUGCGCCACAGUUGUGGCGCUGGGUAUCUGCAAACACACAGAUAA-CAUACA-A .............(((((.(((((.(((....(((.(((((....))))).)))..))).))).)))))))(((((........)))))..-......-. ( -35.60) >consensus CGAUUACUCCAAUACCCAGGCCCAAGGCAAAAUGGGGCAGCAAUAGUUGGGCCACCGUUGUAGCACUGGGUAUCUGCAAAG___UAAAUAA_UAUAUAUG .........((((((((((.((((........))))((.(((((....((....))))))).)).)))))))).))........................ (-19.68 = -20.35 + 0.67)

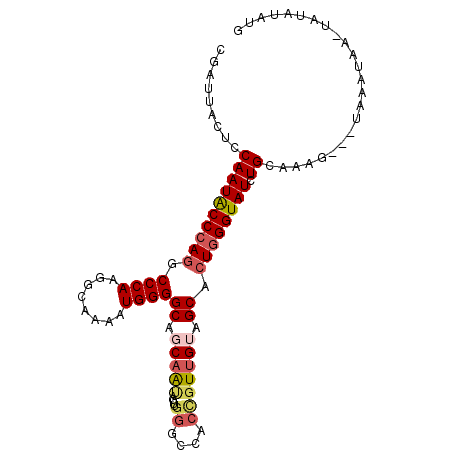
| Location | 19,522,805 – 19,522,901 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -21.97 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19522805 96 - 27905053 CAAAUAUA-UUAUUUA---CUUUGCAGAUACCCAGUGCUACAACGGUGGCCCAACUAUUGCUGCCCCAUUUUGCCUUGGGACUGGGUAUUGGAGUAAUCG ........-....(((---(((....((((((((((((..(((..((......))..)))..))((((........)))))))))))))).))))))... ( -31.00) >DroSec_CAF1 17587 96 - 1 CAUAUAUA-UUAUUUA---CUUUGCAGAUACCCAGUGCUACAACAGUGGCCCAACUAUUGCUGCCCCAUUUUGCCUUGGGACUGGGUAUUGGAGUAAUCG ........-....(((---(((....((((((((((((..(((.(((......))).)))..))((((........)))))))))))))).))))))... ( -30.50) >DroSim_CAF1 17776 96 - 1 CAUAUAUA-UUAUUUA---CUUUGCAGAUACCCAGUGCUACAACAGUGGCCCAACUAUUGCUGCCCCAUUUUGCCUUGGGACUGGGUAUUGGAGUAAUCG ........-....(((---(((....((((((((((((..(((.(((......))).)))..))((((........)))))))))))))).))))))... ( -30.50) >DroEre_CAF1 17713 97 - 1 CAAUUAUAACUCUGAA---UUUUACAGAUACCCAGUGCCACGACGGUUGCCCAGCUAUUGAUGCCCCAUUUCGCCUUGGGCCUGGGCAUUGGUGUAAUCG ..........((((..---.....))))(((((((((((.(((..(((....)))..)))..((((...........))))...)))))))).))).... ( -27.80) >DroYak_CAF1 17797 97 - 1 CAAUUAUAACUCUGAA---CCUUACAGAUACCGAGUGCUACAACGGUUGCCCAACUGCUGAUGCCCCAUUUCGCCUUGGGCCUGGGCAUUGGUGUAAUCG ..........((((..---.....))))...............(((((((((((.((((.(.((((...........)))).).)))))))).))))))) ( -26.40) >DroMoj_CAF1 14855 98 - 1 U-UGUAUG-UUAUCUGUGUGUUUGCAGAUACCCAGCGCCACAACUGUGGCGCAGCUCUUGCUGCCCCAUUUUGCUCUGGGCUUGGGCAUUGGGGUGAUUG .-......-.(((((((......)))))))....(((((((....)))))))(((....)))((((((...(((((.......))))).))))))..... ( -41.70) >consensus CAAAUAUA_UUAUUUA___CUUUGCAGAUACCCAGUGCUACAACGGUGGCCCAACUAUUGCUGCCCCAUUUUGCCUUGGGACUGGGCAUUGGAGUAAUCG ..........................(((((((((((((.((.((((((.....)))))).)).((((........))))....)))))))).))).)). (-21.97 = -21.67 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:04 2006