
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,490,138 – 19,490,257 |
| Length | 119 |
| Max. P | 0.910152 |

| Location | 19,490,138 – 19,490,257 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -0.81 |
| Energy contribution | -0.45 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.04 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
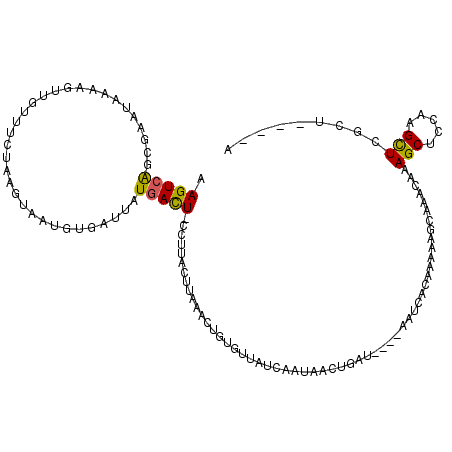
>3R_DroMel_CAF1 19490138 119 + 27905053 AAGUCAGCGAAUAAAAUUUGUUUCAAAGUAUUGUGAUUAUGACUGCCUUACUUAAACUGUGUUAUCAAUAAGUGAUUGACAAUCACGAAAAACAAACAAAGCUCCAAGCUCGCUUAUUA .(((.(((((......(((((((.......((((((((.((((.................))))(((((.....))))).)))))))).....)))))))((.....))))))).))). ( -20.13) >DroSec_CAF1 89017 109 + 1 AAGUCAGCGAAUAAGAGUUGUUUCUAAGUAAUGUGAUUAUGACU-CCUUACUUAAACUGUGUUAUCAAUAAGUGAU-----AACACAAAUAUCAAACAAAGCCCCAAGCUCGUU----A .....(((((.......((((((.(((((((.(.((.......)-)))))))))...((((((((((.....))))-----))))))......)))))).((.....)))))))----. ( -22.40) >DroSim_CAF1 98721 109 + 1 AAGUCAGCGAAUAAAAGUUGUUUCUAAGUAUUGUGGUUAUGAUU-CCUUACUUAAGCUGUGUUAUCAAUAACUGAU-----AACACAAAUAUCAAACAAAGCCCCAAGCUCGCU----A .....(((((.......((((((.((((((....((........-)).))))))...((((((((((.....))))-----))))))......)))))).((.....)))))))----. ( -26.10) >DroEre_CAF1 90098 114 + 1 AAGUCGUUCAAUGAAAGUUGUUUUUAAGUCAUAUUAUUAUGACU-UCUUACUUAAAAUAUCGUAGCAAUAACUUAUAUUUAAUCACUAUAAGCAAACAAAGCUCGAAGUUUACU----U ((((....((((....)))).....((((((((....)))))))-)...)))).((((.(((.(((.....((((((.........))))))........)))))).))))...----. ( -17.52) >DroYak_CAF1 101881 114 + 1 AAGUGACUCAAUGAAAAUAGUUUCUAAAUCAUAAUAUUAUGACU-CCUUACACAAAAUAUAGUAACAAUAAAUUUUCUUUAAUCACUAUAAGCAAACAAAGCUCCAAGUUUGCU----A .(((((......((((((..........(((((....)))))..-..((((..........))))......)))))).....)))))...(((((((..........)))))))----. ( -16.80) >consensus AAGUCAGCGAAUAAAAGUUGUUUCUAAGUAAUGUGAUUAUGACU_CCUUACUUAAACUGUGUUAUCAAUAACUGAU____AAUCACAAAAAGCAAACAAAGCUCCAAGCUCGCU____A .(((((.................................))))).......................................................(((.....)))......... ( -0.81 = -0.45 + -0.36)

| Location | 19,490,138 – 19,490,257 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -4.97 |
| Energy contribution | -5.57 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19490138 119 - 27905053 UAAUAAGCGAGCUUGGAGCUUUGUUUGUUUUUCGUGAUUGUCAAUCACUUAUUGAUAACACAGUUUAAGUAAGGCAGUCAUAAUCACAAUACUUUGAAACAAAUUUUAUUCGCUGACUU .....((((((..(((((.(((((((.......(((((((((....((((((((......))))..))))..))))))))).....(((....)))))))))))))))))))))..... ( -24.60) >DroSec_CAF1 89017 109 - 1 U----AACGAGCUUGGGGCUUUGUUUGAUAUUUGUGUU-----AUCACUUAUUGAUAACACAGUUUAAGUAAGG-AGUCAUAAUCACAUUACUUAGAAACAACUCUUAUUCGCUGACUU .----..((((...((((..((((((.....(((((((-----((((.....))))))))))).((((((((.(-(.......))...))))))))))))))))))..))))....... ( -26.80) >DroSim_CAF1 98721 109 - 1 U----AGCGAGCUUGGGGCUUUGUUUGAUAUUUGUGUU-----AUCAGUUAUUGAUAACACAGCUUAAGUAAGG-AAUCAUAACCACAAUACUUAGAAACAACUUUUAUUCGCUGACUU (----((((((...((((..((((((.....(((((((-----(((((...)))))))))))).(((((((.((-........))....)))))))))))))))))..))))))).... ( -27.20) >DroEre_CAF1 90098 114 - 1 A----AGUAAACUUCGAGCUUUGUUUGCUUAUAGUGAUUAAAUAUAAGUUAUUGCUACGAUAUUUUAAGUAAGA-AGUCAUAAUAAUAUGACUUAAAAACAACUUUCAUUGAACGACUU (----(((((((..........)))))))).((((((..((((((.(((....)))...))))))...((...(-(((((((....))))))))....)).....))))))........ ( -19.80) >DroYak_CAF1 101881 114 - 1 U----AGCAAACUUGGAGCUUUGUUUGCUUAUAGUGAUUAAAGAAAAUUUAUUGUUACUAUAUUUUGUGUAAGG-AGUCAUAAUAUUAUGAUUUAGAAACUAUUUUCAUUGAGUCACUU .----(((((((..........)))))))...(((((((...((((((......((((..........)))).(-(((((((....)))))))).......))))))....))))))). ( -22.50) >consensus U____AGCGAGCUUGGAGCUUUGUUUGAUUUUAGUGAUU____AUCACUUAUUGAUAACACAGUUUAAGUAAGG_AGUCAUAAUCACAUUACUUAGAAACAACUUUUAUUCGCUGACUU .....(((((((..........)))))))..................................(((((((((................)))))))))...................... ( -4.97 = -5.57 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:57 2006