
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
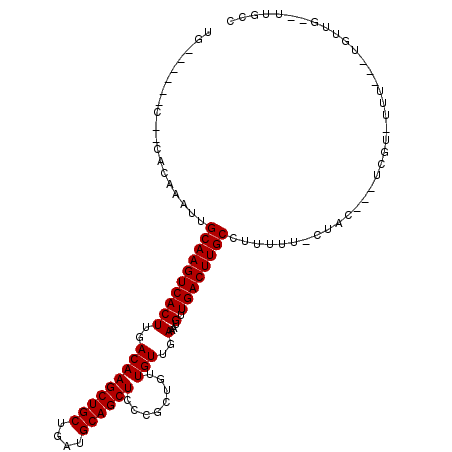
| Location | 2,623,997 – 2,624,096 |
| Length | 99 |
| Max. P | 0.755818 |

| Location | 2,623,997 – 2,624,096 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
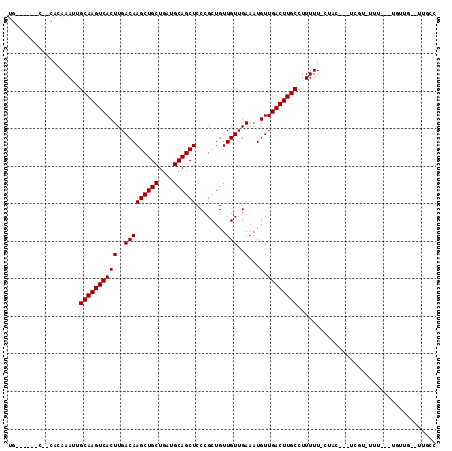
>3R_DroMel_CAF1 2623997 99 + 27905053 UG------C--CACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUU-CCUUUGCUCGU-UGU---UAUUG--UUGCC .(------(--.((((...((((((((((..(((((((((....))))))........)))..)...).))))))))......-.....((....-.))---..)))--).)). ( -24.70) >DroPse_CAF1 19939 107 + 1 UGGCUCUGCCACACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUU-CUAC---UCUUUUUU---UGUUGCCUUGCC .(((...((.(((.(((..((((((((((..(((((((((....))))))........)))..)...).))))))))......-....---....))).---))).))...))) ( -26.99) >DroWil_CAF1 44613 100 + 1 UG------C--CACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUU-CACC---CCGUGUCCCCCCCUCU--AUGCC ..------.--((((((..((((((((((..(((((((((....))))))........)))..)...).))))))))..))).-....---..)))...........--..... ( -21.81) >DroYak_CAF1 6116 96 + 1 UG------C--CACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUU-CCCCAUCU----UGU---UGUUG--UUGCC .(------(--.((((...((((((((((..(((((((((....))))))........)))..)...).))))))))......-...((...----...---)))))--).)). ( -24.60) >DroAna_CAF1 33150 100 + 1 UG------C--CACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUUUCUUCAUCGUGC-CUU---UGUUG--UUGCC .(------(--.((((...((((((((((..(((((((((....))))))........)))..)...).)))))))).............((...-...---)))))--).)). ( -23.70) >DroPer_CAF1 19956 105 + 1 UGGCUCUGCCACACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUU-CUAC---UCUUUUUU-----UUGCCUUGCC ((((...))))........((((((((((..(((((((((....))))))........)))..)...).))))))))......-....---........-----.......... ( -24.80) >consensus UG______C__CACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGUUGACUUGCCUUUUU_CUAC___UCGU_UUU___UGUUG__UUGCC ...................((((((((((..(((((((((....))))))........)))..)...).))))))))..................................... (-21.70 = -21.70 + -0.00)

| Location | 2,623,997 – 2,624,096 |
|---|---|
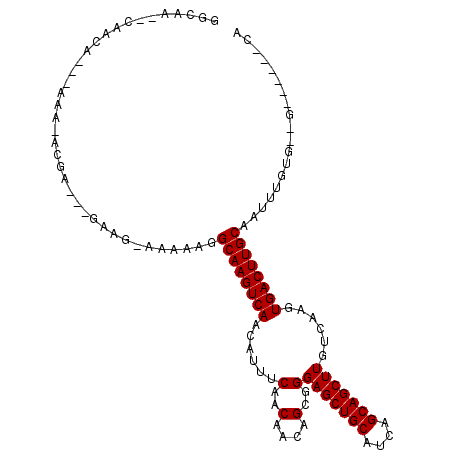
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
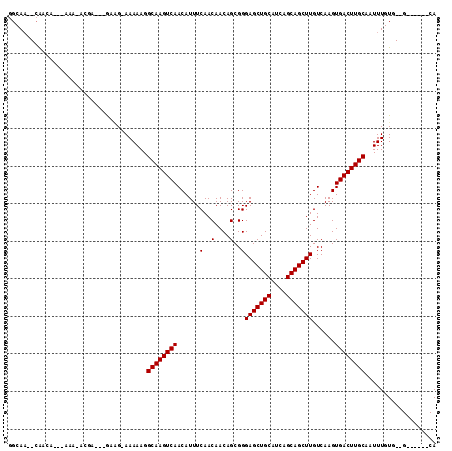
>3R_DroMel_CAF1 2623997 99 - 27905053 GGCAA--CAAUA---ACA-ACGAGCAAAGG-AAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUG--G------CA (....--)....---...-....((...(.-(((...((((((((......(..(....)..)(((((((....)))))))......))))))))..))).).--)------). ( -25.40) >DroPse_CAF1 19939 107 - 1 GGCAAGGCAACA---AAAAAAGA---GUAG-AAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUGUGGCAGAGCCA (((...((.(((---((......---....-......((((((((......(..(....)..)(((((((....)))))))......))))))))..)))))...))...))). ( -30.07) >DroWil_CAF1 44613 100 - 1 GGCAU--AGAGGGGGGGACACGG---GGUG-AAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUG--G------CA (.(((--(((...(....)....---....-......((((((((......(..(....)..)(((((((....)))))))......))))))))..))))))--.------). ( -29.40) >DroYak_CAF1 6116 96 - 1 GGCAA--CAACA---ACA----AGAUGGGG-AAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUG--G------CA .((.(--((((.---.(.----....)..)-......((((((((......(..(....)..)(((((((....)))))))......))))))))...)))).--)------). ( -25.90) >DroAna_CAF1 33150 100 - 1 GGCAA--CAACA---AAG-GCACGAUGAAGAAAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUG--G------CA (....--)....---..(-.(((((............((((((((......(..(....)..)(((((((....)))))))......))))))))...)))))--.------). ( -27.16) >DroPer_CAF1 19956 105 - 1 GGCAAGGCAA-----AAAAAAGA---GUAG-AAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUGUGGCAGAGCCA .....(((..-----........---....-......((((((((......(..(....)..)(((((((....)))))))......))))))))..((((.....))))))). ( -28.80) >consensus GGCAA__CAACA___AAA_ACGA___GAAG_AAAAAGGCAAGUCAACAUUUCAACAACAGCGGGAGCUGCAUCAGCAGCUUGUCAAGUGACUUGCAAUUUGUG__G______CA .....................................((((((((......(..(....)..)(((((((....)))))))......))))))))................... (-21.70 = -21.70 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:40 2006