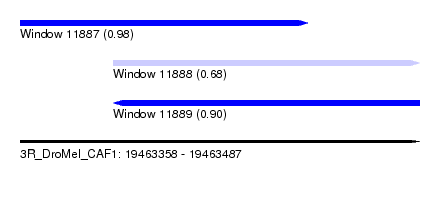
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,463,358 – 19,463,487 |
| Length | 129 |
| Max. P | 0.984787 |

| Location | 19,463,358 – 19,463,451 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -24.69 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19463358 93 + 27905053 ACAGAUCGGGAGUUGUAGUCAAAAUGGGGG------------------------GCCGAGUUGGCUUGGAGCUUGGAGCUUCAGGCUUGGGGCUGGGCCUGUUGAUGGGGCUUCAAU ........((((((...(((((......((------------------------(((.((((((((((((((.....))))))))))...)))).))))).)))))..))))))... ( -37.50) >DroSec_CAF1 62432 77 + 1 GCAGAUCGGGAGUUGUAGUCAAAAUGGG-G------------------------GCCGAGU-GGCUUGGAGCU--------------UGGGGCUGGGCCUGUUGAUGGGGCUUCAAU ........((((((...(((((.....(-(------------------------(((.(((-(((.....)))--------------....))).))))).)))))..))))))... ( -24.20) >DroSim_CAF1 71487 92 + 1 GCAGAUCGGGAGUUGUAGUCAAAAUGGGGG------------------------GCCGAGU-GGCUUGGAGCUUGGAGCUUCAGGCUUGGGGCUGGGCCUGUUGAUGGGGCUUCAAU ........((((((...(((((......((------------------------(((.(((-((((((((((.....))))))))))....))).))))).)))))..))))))... ( -36.60) >DroEre_CAF1 61864 84 + 1 GCAGAUCGGGAGUUGUAGUCAAAUUGGGG-------------------------GCCAAGU-GGCUUGGAGCUU-------GGGGCUUGGGGCUGGGUCUGUUGAUGGGGCUUCAAU ...........((((.((((..((..(((-------------------------(((....-(((((.((((..-------...)))).))))).))))).)..))..)))).)))) ( -27.90) >DroYak_CAF1 73564 116 + 1 GCAAAUCGGGAGUUGUAGUCAAAAUGGGGGGCUAUGGAGCCAUGGAGCCAUGGAGCCAAGU-GGCUUGGAGCUUGAAGCUGGGGGCUUGGGGCUGGGUCUGUUGAUGGGGCUUCAAU ........((..(..(((((.........)))))..)..)).(((((((.(.(((((....-))))).)(((((.((((.....)))).)))))..(((....)))..))))))).. ( -41.20) >consensus GCAGAUCGGGAGUUGUAGUCAAAAUGGGGG________________________GCCGAGU_GGCUUGGAGCUUG_AGCU_CAGGCUUGGGGCUGGGCCUGUUGAUGGGGCUUCAAU ........((((((...(((((...(((...........................(((((....)))))(((((.((((.....)))).)))))...))).)))))..))))))... (-24.69 = -25.48 + 0.79)

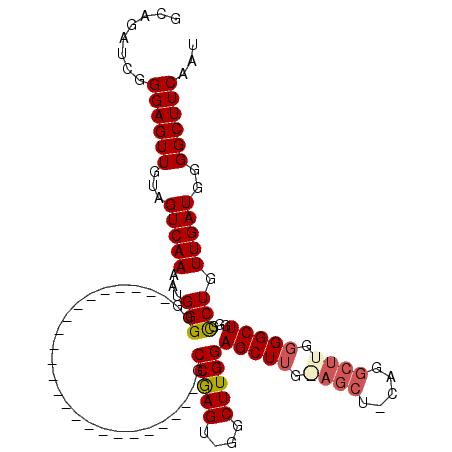
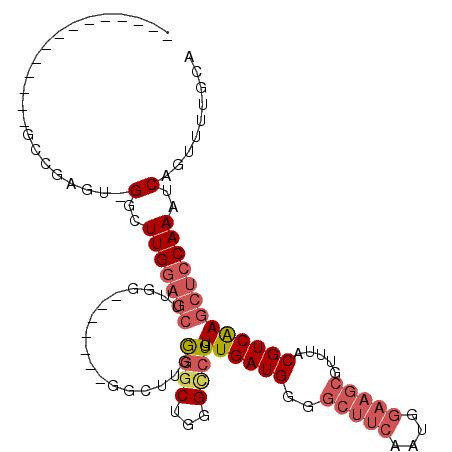
| Location | 19,463,388 – 19,463,487 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -18.96 |
| Energy contribution | -20.35 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19463388 99 + 27905053 --------------GCCGAGUUGGCUUGGAGCUUGGAGCUUCAGGCUUGGGGCUGGGCCUGUUGAUGGGGCUUCAAUGGAAGCGUUUACGUCAAGCUCCAAAUCAGUUUUGCA --------------((.((..((..((((((((((..(...(((((((......)))))))........(((((....))))).....)..))))))))))..))..)).)). ( -41.00) >DroSec_CAF1 62461 84 + 1 --------------GCCGAGU-GGCUUGGAGCU--------------UGGGGCUGGGCCUGUUGAUGGGGCUUCAAUGAAAGCGUUUACGUCAAGCUCCAAAUCAGUUUUGCA --------------((.((.(-(..((((((((--------------(((.(.((((((((((((.......)))))...)).)))))).)))))))))))..)).))..)). ( -30.80) >DroSim_CAF1 71517 98 + 1 --------------GCCGAGU-GGCUUGGAGCUUGGAGCUUCAGGCUUGGGGCUGGGCCUGUUGAUGGGGCUUCAAUGGAAGCGUUUACGUCAAGCUCCAAAUCAGUUUUGCA --------------((.((.(-(..((((((((((..(...(((((((......)))))))........(((((....))))).....)..))))))))))..)).))..)). ( -39.40) >DroEre_CAF1 61893 91 + 1 --------------GCCAAGU-GGCUUGGAGCUU-------GGGGCUUGGGGCUGGGUCUGUUGAUGGGGCUUCAAUGGAAGCGUUUACGUCAAGCUCCAAAUCAGUUUUGCA --------------((.((.(-(..(((((((((-------((.(...((((((..(((....)))..))))))((((....))))..).)))))))))))..)).))..)). ( -35.30) >DroYak_CAF1 73604 112 + 1 CAUGGAGCCAUGGAGCCAAGU-GGCUUGGAGCUUGAAGCUGGGGGCUUGGGGCUGGGUCUGUUGAUGGGGCUUCAAUGGAAGCGUUUACGUCAAGCUGCAAAUCAGUUUUGCA ...(((.((.(.(((((....-))))).)(((((.((((.....)))).))))))).))).((((((..(((((....))))).....))))))...(((((.....))))). ( -40.50) >DroAna_CAF1 82625 78 + 1 ----------------------GUCUUGGAAACUGG------------GAGGCUGGGAGUGUUGAUGGGGCACCAAUGGAACAGUUAACGUCGAGCGCCAAAUCA-UUUUGCU ----------------------((((((....)...------------)))))....((((.((((..(((.((.(((..........))).).).)))..))))-...)))) ( -16.00) >consensus ______________GCCGAGU_GGCUUGGAGCUUGG_______GGCUUGGGGCUGGGCCUGUUGAUGGGGCUUCAAUGGAAGCGUUUACGUCAAGCUCCAAAUCAGUUUUGCA ......................(..(((((((.................((((...)))).((((((..(((((....))))).....)))))))))))))..)......... (-18.96 = -20.35 + 1.39)
| Location | 19,463,388 – 19,463,487 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.93 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19463388 99 - 27905053 UGCAAAACUGAUUUGGAGCUUGACGUAAACGCUUCCAUUGAAGCCCCAUCAACAGGCCCAGCCCCAAGCCUGAAGCUCCAAGCUCCAAGCCAACUCGGC-------------- ............((((((((((..((....(((((....)))))........(((((..........)))))..))..))))))))))(((.....)))-------------- ( -32.00) >DroSec_CAF1 62461 84 - 1 UGCAAAACUGAUUUGGAGCUUGACGUAAACGCUUUCAUUGAAGCCCCAUCAACAGGCCCAGCCCCA--------------AGCUCCAAGCC-ACUCGGC-------------- ............((((((((((........(((((....)))))..........(((...))).))--------------))))))))(((-....)))-------------- ( -24.60) >DroSim_CAF1 71517 98 - 1 UGCAAAACUGAUUUGGAGCUUGACGUAAACGCUUCCAUUGAAGCCCCAUCAACAGGCCCAGCCCCAAGCCUGAAGCUCCAAGCUCCAAGCC-ACUCGGC-------------- ............((((((((((..((....(((((....)))))........(((((..........)))))..))..))))))))))(((-....)))-------------- ( -32.00) >DroEre_CAF1 61893 91 - 1 UGCAAAACUGAUUUGGAGCUUGACGUAAACGCUUCCAUUGAAGCCCCAUCAACAGACCCAGCCCCAAGCCCC-------AAGCUCCAAGCC-ACUUGGC-------------- ............((((((((((........(((((....)))))................((.....))..)-------)))))))))(((-....)))-------------- ( -24.60) >DroYak_CAF1 73604 112 - 1 UGCAAAACUGAUUUGCAGCUUGACGUAAACGCUUCCAUUGAAGCCCCAUCAACAGACCCAGCCCCAAGCCCCCAGCUUCAAGCUCCAAGCC-ACUUGGCUCCAUGGCUCCAUG ((((((.....))))))((((((.((....(((((....)))))................((.....)).....)).))))))....((((-....)))).((((....)))) ( -26.70) >DroAna_CAF1 82625 78 - 1 AGCAAAA-UGAUUUGGCGCUCGACGUUAACUGUUCCAUUGGUGCCCCAUCAACACUCCCAGCCUC------------CCAGUUUCCAAGAC---------------------- .......-((((..((((((.((((.....)))).....))))))..))))..............------------..............---------------------- ( -11.00) >consensus UGCAAAACUGAUUUGGAGCUUGACGUAAACGCUUCCAUUGAAGCCCCAUCAACAGGCCCAGCCCCAAGCC_______CCAAGCUCCAAGCC_ACUCGGC______________ ...........(((((((((((..(....)(((((....)))))..................................)))))))))))........................ (-12.82 = -13.93 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:49 2006