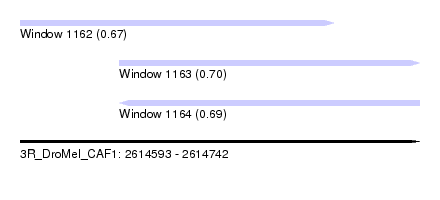
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,614,593 – 2,614,742 |
| Length | 149 |
| Max. P | 0.697937 |

| Location | 2,614,593 – 2,614,710 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.54 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -21.22 |
| Energy contribution | -23.22 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2614593 117 + 27905053 CAGACUGCUUCAGACUAUUUCCUAUAUAAAAUAAUCAAAUUACAACACUCUGCAGUGGUAUCAAACACGCUGGGCGUCUUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCAGCAU ......(((..(((((((((........)))))..............(((.((.(((........))))).))).)))).)))((((((.((((((......)))))).)))))).. ( -28.70) >DroSec_CAF1 14365 115 + 1 CAGACGAUUUCU--UUAGCAGCUAUGUGAAAAAAUCAAAUUACAACACUCAGCAGUGGUAUCAAAACCGCUGGGAGUCAUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCCGCAU ............--...((.(((((((((.....))).........((((..(((((((......))))))).))))))))))...(((.((((((......)))))).))).)).. ( -32.90) >DroSim_CAF1 2463 115 + 1 CAGACUAUUUCU--UUAGCAGCUAUGUGAAAAAAUCAAAUUACAACACUCAGCAGUGGUAUCAAAACCGCUGGGAGUCAUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCCGCAU ..(.(((.....--.)))).(((((((((.....))).........((((..(((((((......))))))).))))))))))((.(((.((((((......)))))).))).)).. ( -33.40) >consensus CAGACUAUUUCU__UUAGCAGCUAUGUGAAAAAAUCAAAUUACAACACUCAGCAGUGGUAUCAAAACCGCUGGGAGUCAUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCCGCAU ....................((((((....................((((..(((((((......))))))).))))))))))((.(((.((((((......)))))).))).)).. (-21.22 = -23.22 + 2.00)

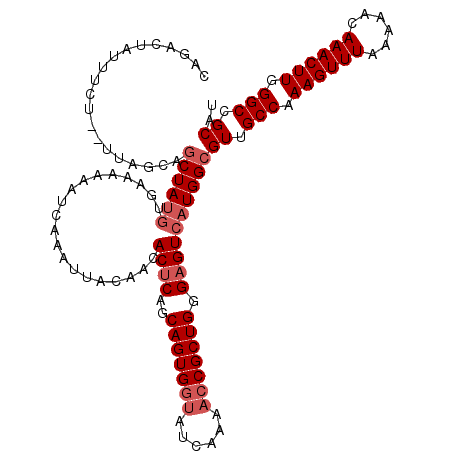
| Location | 2,614,630 – 2,614,742 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -24.81 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2614630 112 + 27905053 AAUUACAACACUCUGCAGUGGUAUCAAACACGCUGGGCGUCUUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCAGCAUGUUUUUUUUAUGUGUUGCUAAAAUUUCCAUGU-------- ..(((((((((......(((........)))((..((...))..))((((((.((((((......)))))).)))))).............)))))).)))...........-------- ( -29.90) >DroSec_CAF1 14400 119 + 1 AAUUACAACACUCAGCAGUGGUAUCAAAACCGCUGGGAGUCAUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCCGCAUGUUAU-GUUAUGUGUUUCCAAAAUCUCCAUAUUGAAUCUG ...........(((((((((((......)))))))((((...(((....(((.((((((......)))))).)))((((((....-..))))))...)))....))))...))))..... ( -32.50) >DroSim_CAF1 2498 119 + 1 AAUUACAACACUCAGCAGUGGUAUCAAAACCGCUGGGAGUCAUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCCGCAUGUUAU-GUUAUGUGUUUCCAAAAUCUCCAUAUUGAAUCUG ...........(((((((((((......)))))))((((...(((....(((.((((((......)))))).)))((((((....-..))))))...)))....))))...))))..... ( -32.50) >consensus AAUUACAACACUCAGCAGUGGUAUCAAAACCGCUGGGAGUCAUGGCGUUGCCAAAGUUUAAAAACAAACUUGGGCCGCAUGUUAU_GUUAUGUGUUUCCAAAAUCUCCAUAUUGAAUCUG ...............(((((((......)))))))((((...(((....(((.((((((......)))))).)))((((((.......))))))...)))....))))............ (-24.81 = -25.70 + 0.89)

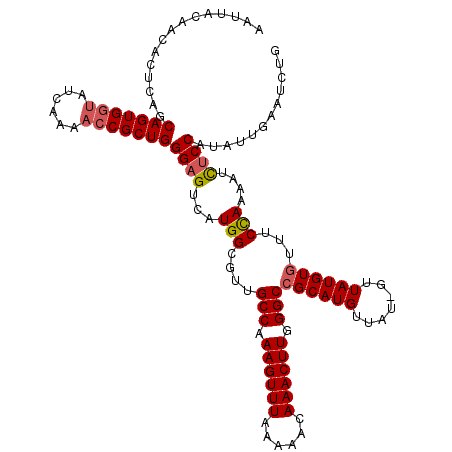
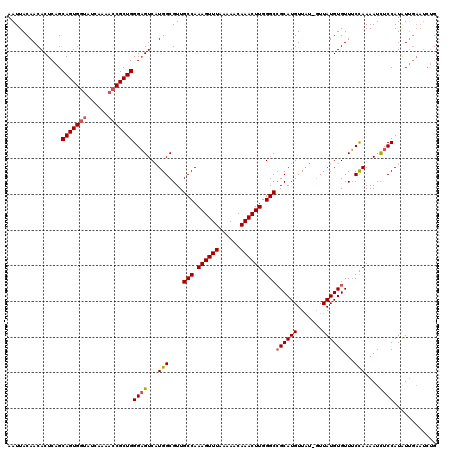
| Location | 2,614,630 – 2,614,742 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -22.23 |
| Energy contribution | -23.90 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2614630 112 - 27905053 --------ACAUGGAAAUUUUAGCAACACAUAAAAAAAACAUGCUGCCCAAGUUUGUUUUUAAACUUUGGCAACGCCAAGACGCCCAGCGUGUUUGAUACCACUGCAGAGUGUUGUAAUU --------..............(((((((.......((((((((((((.(((((((....))))))).)))...((......))..))))))))).......(....).))))))).... ( -29.10) >DroSec_CAF1 14400 119 - 1 CAGAUUCAAUAUGGAGAUUUUGGAAACACAUAAC-AUAACAUGCGGCCCAAGUUUGUUUUUAAACUUUGGCAACGCCAUGACUCCCAGCGGUUUUGAUACCACUGCUGAGUGUUGUAAUU ..((((((((((((((....((....))......-....(((((((((.(((((((....))))))).)))..)).)))).))))(((((((.........))))))).)))))).)))) ( -35.70) >DroSim_CAF1 2498 119 - 1 CAGAUUCAAUAUGGAGAUUUUGGAAACACAUAAC-AUAACAUGCGGCCCAAGUUUGUUUUUAAACUUUGGCAACGCCAUGACUCCCAGCGGUUUUGAUACCACUGCUGAGUGUUGUAAUU ..((((((((((((((....((....))......-....(((((((((.(((((((....))))))).)))..)).)))).))))(((((((.........))))))).)))))).)))) ( -35.70) >consensus CAGAUUCAAUAUGGAGAUUUUGGAAACACAUAAC_AUAACAUGCGGCCCAAGUUUGUUUUUAAACUUUGGCAACGCCAUGACUCCCAGCGGUUUUGAUACCACUGCUGAGUGUUGUAAUU ........................(((((..........(((((((((.(((((((....))))))).)))..)).)))).....(((((((.........))))))).)))))...... (-22.23 = -23.90 + 1.67)

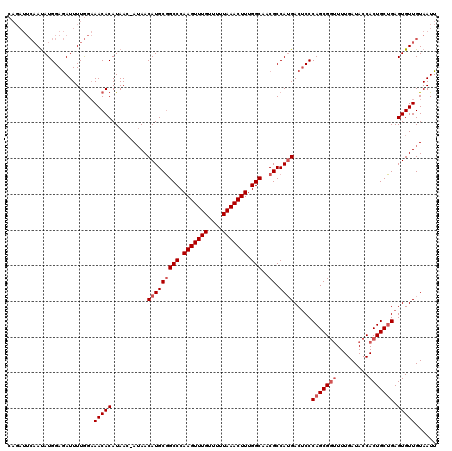
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:38 2006