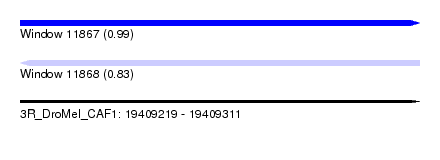
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,409,219 – 19,409,311 |
| Length | 92 |
| Max. P | 0.986893 |

| Location | 19,409,219 – 19,409,311 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.97 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
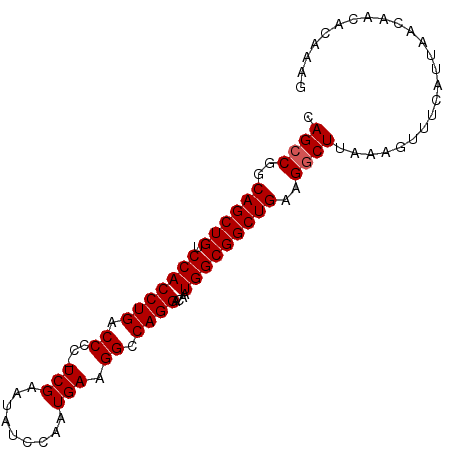
>3R_DroMel_CAF1 19409219 92 + 27905053 CAGCCGGCAGCUGUCCACCUGACCCC-UCGAAUAUCCAAUGAAGGCCAGGACAAUGGCGGCUGAAGGCUUAAAGUUUCAUUAACAACACAAAG .((((..((((((.(((((((.((..-(((.........))).)).))))....)))))))))..))))....(((.....)))......... ( -26.50) >DroSec_CAF1 10067 92 + 1 CAGCCGGCAGCUGUCCACCUGACCCC-UCGAAUAUCCAAUGAAGGCCAGGACAAUGGCGGCUGACGGCUUAAAGUUUCAUUAAAAACACAAAG .(((((.((((((.(((((((.((..-(((.........))).)).))))....))))))))).)))))....((((......))))...... ( -28.50) >DroSim_CAF1 10816 92 + 1 CAGCCGGCAGCUGUCCACCUGACCCC-UCGAAUAUCCAAUGAAGGCCAGGACAAUGGCGGCUGAAGGCUUAAAGUUUCAUUAACAACACAAAG .((((..((((((.(((((((.((..-(((.........))).)).))))....)))))))))..))))....(((.....)))......... ( -26.50) >DroEre_CAF1 9955 92 + 1 CAGACGGCAGCUGUCCACCUGACCCC-UCGAAUGGCCAAUGAAGGCCAGGACAAUGGCGGCUGAAGGCUUAAAGUUUCAUUAACAACACAAAG (((.((.((..(((((...(((....-)))..(((((......)))))))))).)).)).)))..........(((.....)))......... ( -24.50) >DroYak_CAF1 10427 93 + 1 CAGCCGCCAGCUGUCCACCUGACCCCUUCGAAUAUCCAAUGAAGGCCAGGACAAUGGCGGCUGAAGGCUUAAAGUUUCAUUAACAACACAAAG (((((((((..(((((...((...((((((.........)))))).))))))).)))))))))..........(((.....)))......... ( -30.50) >consensus CAGCCGGCAGCUGUCCACCUGACCCC_UCGAAUAUCCAAUGAAGGCCAGGACAAUGGCGGCUGAAGGCUUAAAGUUUCAUUAACAACACAAAG .((((..((((((.(((((((.((...(((.........))).)).))))....)))))))))..))))........................ (-23.68 = -23.88 + 0.20)


| Location | 19,409,219 – 19,409,311 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.97 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
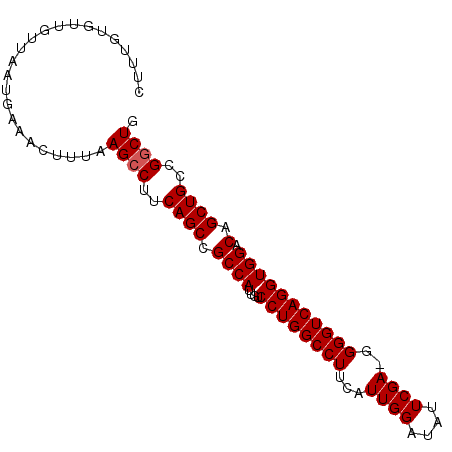
>3R_DroMel_CAF1 19409219 92 - 27905053 CUUUGUGUUGUUAAUGAAACUUUAAGCCUUCAGCCGCCAUUGUCCUGGCCUUCAUUGGAUAUUCGA-GGGGUCAGGUGGACAGCUGCCGGCUG ......(((........)))....((((..((((.((((....((((((((...((((....))))-.))))))))))).).))))..)))). ( -29.10) >DroSec_CAF1 10067 92 - 1 CUUUGUGUUUUUAAUGAAACUUUAAGCCGUCAGCCGCCAUUGUCCUGGCCUUCAUUGGAUAUUCGA-GGGGUCAGGUGGACAGCUGCCGGCUG ......(((((....)))))....(((((.((((.((((....((((((((...((((....))))-.))))))))))).).)))).))))). ( -33.90) >DroSim_CAF1 10816 92 - 1 CUUUGUGUUGUUAAUGAAACUUUAAGCCUUCAGCCGCCAUUGUCCUGGCCUUCAUUGGAUAUUCGA-GGGGUCAGGUGGACAGCUGCCGGCUG ......(((........)))....((((..((((.((((....((((((((...((((....))))-.))))))))))).).))))..)))). ( -29.10) >DroEre_CAF1 9955 92 - 1 CUUUGUGUUGUUAAUGAAACUUUAAGCCUUCAGCCGCCAUUGUCCUGGCCUUCAUUGGCCAUUCGA-GGGGUCAGGUGGACAGCUGCCGUCUG ......((((((.............((((...(((.((.(((...(((((......)))))..)))-))))).)))).))))))......... ( -24.64) >DroYak_CAF1 10427 93 - 1 CUUUGUGUUGUUAAUGAAACUUUAAGCCUUCAGCCGCCAUUGUCCUGGCCUUCAUUGGAUAUUCGAAGGGGUCAGGUGGACAGCUGGCGGCUG (((...(((........)))...)))....((((((((((((((((((((((..((((....)))).)))))))...)))))).))))))))) ( -35.00) >consensus CUUUGUGUUGUUAAUGAAACUUUAAGCCUUCAGCCGCCAUUGUCCUGGCCUUCAUUGGAUAUUCGA_GGGGUCAGGUGGACAGCUGCCGGCUG ........................((((..((((.((((....((((((((...((((....))))..))))))))))).).))))..)))). (-27.40 = -27.60 + 0.20)

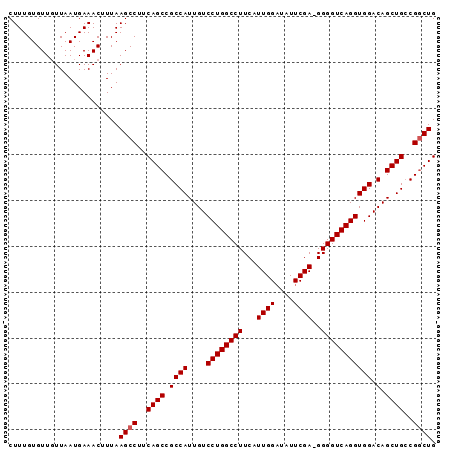
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:29 2006