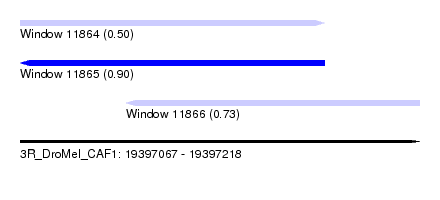
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,397,067 – 19,397,218 |
| Length | 151 |
| Max. P | 0.902797 |

| Location | 19,397,067 – 19,397,182 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.84 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19397067 115 + 27905053 ACACACAGCGUUGCAUUUAGACAAUGCUGAGAUGCUGUGCUUGUGGCAUGUUUACAAGUUGAAUG-----UGUAUUUUUCCACAAAACUCAUUGAAAUGGAAUUCCAUUCGGAGCACAUC ...((((((((((((((.....)))))...)))))))))..(((((((((((((.....))))))-----)))....((((.(((......))).(((((....))))).)))))))).. ( -31.50) >DroSec_CAF1 45194 115 + 1 ACACACAGCGUUGCAUUUAGACAAUGCUGAGAUGCUGGGCUUGUGGCAUAUUUACGAGUUGAAUG-----UUUAUUUUUCCACAAAACUCAUUGAAAUGGAAUUCCAUUCGGAGCACAUC .....((((((((........))))))))...((((.(((((((((.....)))))))))(((((-----.......((((((((......)))...)))))...)))))..)))).... ( -27.70) >DroEre_CAF1 49823 115 + 1 ACACACAGCGUUGCAUUUAGACAAAGCUGAGAUGCUGUGCUUGUGGCAUAUUUACGAGUUGUAUG-----UGUAUUUUUGCACAAAACUCAUUGAAAUGGAAUUCCAUUCGGAGCACAUC .((((.((((..((((((((......)).))))))..))))))))((........(((((...((-----((((....)))))).))))).(((((.(((....)))))))).))..... ( -35.00) >DroYak_CAF1 50131 115 + 1 ACACACAGCGUUGCAUUUAGACAAUGCUGAGAUGCUGUGCUUGUGGCAUAUUUACGAGUUGUAUG-----UGUGUUUUUCCACAAAACUCAUUGAAAUGGAAUUCCAUUCGGAGCACAUC .....((((((((........))))))))......((((((((((((((((.(((.....))).)-----)))).....)))).........((((.(((....)))))))))))))).. ( -32.80) >DroAna_CAF1 52786 116 + 1 GCAUCCA---AUGUACUUAGACAAUACCGAGAUGCCGCACUGUGGAAAUAUUUACAUUUCGUGUGUAUCGUGUGCUGUUUCAG-AUAUUCAUUGAAAUGGAAUCCCAUUCGAGGCACAUC ((.((((---(((..((.(((((.(((...(((((.((((((((((....))))))....)))))))))..))).))))).))-.....))))).(((((....))))).)).))..... ( -26.70) >consensus ACACACAGCGUUGCAUUUAGACAAUGCUGAGAUGCUGUGCUUGUGGCAUAUUUACGAGUUGUAUG_____UGUAUUUUUCCACAAAACUCAUUGAAAUGGAAUUCCAUUCGGAGCACAUC .....((((((((........))))))))...((((..((((((((.....))))))))................................(((((.(((....)))))))))))).... (-19.56 = -20.84 + 1.28)

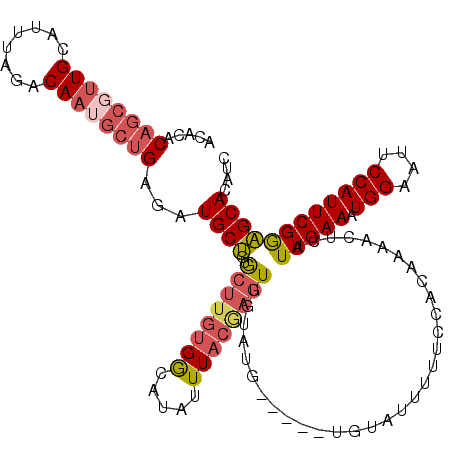

| Location | 19,397,067 – 19,397,182 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -21.78 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19397067 115 - 27905053 GAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAAUACA-----CAUUCAACUUGUAAACAUGCCACAAGCACAGCAUCUCAGCAUUGUCUAAAUGCAACGCUGUGUGU ..((((((..((((((....))))))..(..((((.((((........))-----))...))))..)............))))))((((..((((.((((......)))).)))).)))) ( -31.20) >DroSec_CAF1 45194 115 - 1 GAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAAUAAA-----CAUUCAACUCGUAAAUAUGCCACAAGCCCAGCAUCUCAGCAUUGUCUAAAUGCAACGCUGUGUGU ((((((((..((((((....))))))..(((((((..(((..........-----)))..)))))))............)))...))))).((((.((((......)))).))))..... ( -27.20) >DroEre_CAF1 49823 115 - 1 GAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGCAAAAAUACA-----CAUACAACUCGUAAAUAUGCCACAAGCACAGCAUCUCAGCUUUGUCUAAAUGCAACGCUGUGUGU ..((((((..((((((....))))))..(((((((.((((........))-----))...)))))))............))))))((((..((((.((((......)))).)))).)))) ( -32.60) >DroYak_CAF1 50131 115 - 1 GAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAACACA-----CAUACAACUCGUAAAUAUGCCACAAGCACAGCAUCUCAGCAUUGUCUAAAUGCAACGCUGUGUGU ..((((((..((((((....))))))..(((((((.((((........))-----))...)))))))............))))))((((..((((.((((......)))).)))).)))) ( -33.90) >DroAna_CAF1 52786 116 - 1 GAUGUGCCUCGAAUGGGAUUCCAUUUCAAUGAAUAU-CUGAAACAGCACACGAUACACACGAAAUGUAAAUAUUUCCACAGUGCGGCAUCUCGGUAUUGUCUAAGUACAU---UGGAUGC (((((.....((((((....))))))......))))-).......((((..(.....)..((((((....))))))....)))).((((((..(((((.....)))))..---.)))))) ( -25.00) >consensus GAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAAUACA_____CAUACAACUCGUAAAUAUGCCACAAGCACAGCAUCUCAGCAUUGUCUAAAUGCAACGCUGUGUGU ..(((((...((((((....))))))..(((((((..(((...............)))..))))))).............)))))((((..((((.((((......)))).)))).)))) (-21.78 = -22.06 + 0.28)



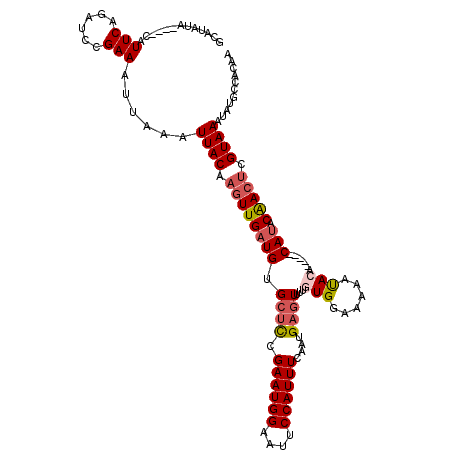
| Location | 19,397,107 – 19,397,218 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.07 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -12.44 |
| Energy contribution | -13.28 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19397107 111 - 27905053 UCGUAUA----CAUUCAGAUCCGAAAUUAAAUUACAAGUUGAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAAUACA-----CAUUCAACUUGUAAACAUGCCACAA ..((((.----..(((......)))......(((((((((((...((((.((((((....))))))....))))..((((........))-----)).)))))))))))..))))..... ( -26.50) >DroSec_CAF1 45234 111 - 1 ACGUAUA----CAUUCAGAUCCGAAAUUAAAUUACAAGUUGAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAAUAAA-----CAUUCAACUCGUAAAUAUGCCACAA ..(((((----..(((......)))......((((.(((((((((((((.((((((....))))))....)))).((((......)))))-----)).)))))).)))).)))))..... ( -21.60) >DroEre_CAF1 49863 111 - 1 GCAUAUA----CAUUCAGAUCCGAAAUUAAAUUACAAGUUGAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGCAAAAAUACA-----CAUACAACUCGUAAAUAUGCCACAA ((((((.----..(((......)))......((((.((((((((((....((((((....))))))......(((((.....))))).))-----))).))))).))))))))))..... ( -24.80) >DroYak_CAF1 50171 115 - 1 GCAUAUAUGUAGAUUCGGAUGCGAAAUUAAAUUACAAGUUGAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAACACA-----CAUACAACUCGUAAAUAUGCCACAA ((((((...(((.((((....)))).)))..((((.(((((((((((((.((((((....))))))....))))...(((......))))-----))).))))).))))))))))..... ( -30.00) >DroAna_CAF1 52823 109 - 1 ------A----CAUUCAAACCCGAAAUUAAAUUACAAGUUGAUGUGCCUCGAAUGGGAUUCCAUUUCAAUGAAUAU-CUGAAACAGCACACGAUACACACGAAAUGUAAAUAUUUCCACA ------.----...........(((((....(((((..(((.((((..(((..(((....)))(((((........-.))))).......)))..)))))))..)))))..))))).... ( -17.50) >consensus GCAUAUA____CAUUCAGAUCCGAAAUUAAAUUACAAGUUGAUGUGCUCCGAAUGGAAUUCCAUUUCAAUGAGUUUUGUGGAAAAAUACA_____CAUACAACUCGUAAAUAUGCCACAA .............(((......)))......((((.((((((((.((((.((((((....))))))....))))...(((......)))......))).))))).))))........... (-12.44 = -13.28 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:27 2006