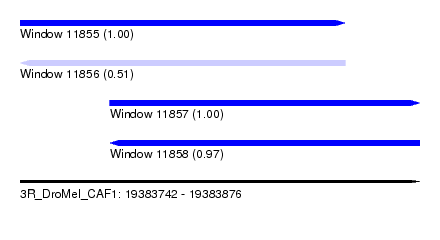
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,383,742 – 19,383,876 |
| Length | 134 |
| Max. P | 0.997190 |

| Location | 19,383,742 – 19,383,851 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.12 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -8.55 |
| Energy contribution | -9.38 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.29 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
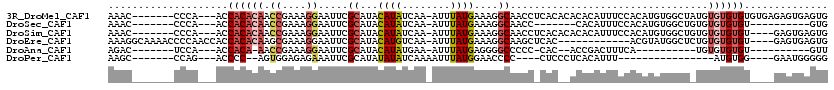
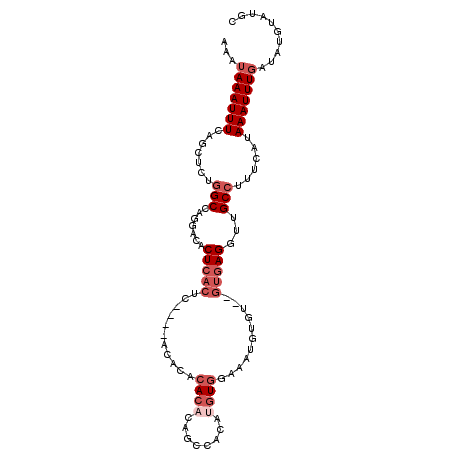
>3R_DroMel_CAF1 19383742 109 + 27905053 AAAC-------CCCA---ACCACACAACCGAAAGGAAUUCGCAUACAUAUCAA-AUUUAUGAAAGGCAACCUCACACACACAUUUCCACAUGUGGCUAUGUGUGUGUGUGAGAGUGAGUG ....-------....---....(((..((....))...((((...((((....-...)))).........(((((((((((((..(((....)))....))))))))))))).))))))) ( -36.90) >DroSec_CAF1 33672 92 + 1 AAAC-------CCCA---ACCACACAACCGAAAGGAAUUCGCAUACAUAUCAA-AUUUAUGAAAGGCAACC-------CACAUUUCCACAUGUGGCUGUGUGUGUGU----------GUG ....-------....---..((((((.((....))....((((((((......-..........(....)(-------(((((......)))))).)))))))))))----------))) ( -26.90) >DroSim_CAF1 39682 105 + 1 AAAC-------CCCA---ACCACACAACCGAAAGGAAUUCGCAUACAUAUCAA-AUUUAUGAAAGGCAACCUCACACACACAUUUCCACAUGUGGCUGUGUGUGUGU----GAGUGAGUG ....-------....---....(((..((....))...((((...((((....-...))))....))...(((((((((((((..(((....)))..))))))))))----))).))))) ( -36.40) >DroEre_CAF1 38147 103 + 1 AAAGGCAAAACCCCAACCACCACACAAGCGAAAGGAAUUCGCAUACAUGUCAA-AUUUAUGAAAGGCAAGCUCAC------------ACGUAUGGCUCUGUGUGUGU----GAGUGAGUG ...............((((((((((..(((((.....)))))(((((((((..-..........))))(((.((.------------.....))))).)))))))))----).))).)). ( -23.40) >DroAna_CAF1 41376 85 + 1 AGAC-------UCCA---ACCACA-AACCGAAAGGAAUUCGCAUACAUAUGAA-AUUUAUGAGGGGCCCCC-CAC--ACCGACUUUCA----------UGUGUGUGU----------GUU ....-------....---......-..((....))....((((((((((((((-(.((.((.(((....))-).)--)..)).)))))----------)))))))))----------).. ( -29.70) >DroPer_CAF1 38467 84 + 1 AAGC-------CCAG---ACCCC--AGUGGAGAGAAAUUCGCAUAUAUAUCAAAAUUUAUGGAACCCC----CUCCCUCACAUUU----------------AUGUGG----GAAUGGGGG ....-------....---.((((--(..((((.(...(((.((((.((......)).)))))))...)----))))((((((...----------------.)))))----)..))))). ( -23.30) >consensus AAAC_______CCCA___ACCACACAACCGAAAGGAAUUCGCAUACAUAUCAA_AUUUAUGAAAGGCAACC_CAC__ACACAUUUCCACAUGUGGCU_UGUGUGUGU____GAGUGAGUG ....................((((((.((....)).....((...((((........))))....)).................................)))))).............. ( -8.55 = -9.38 + 0.84)
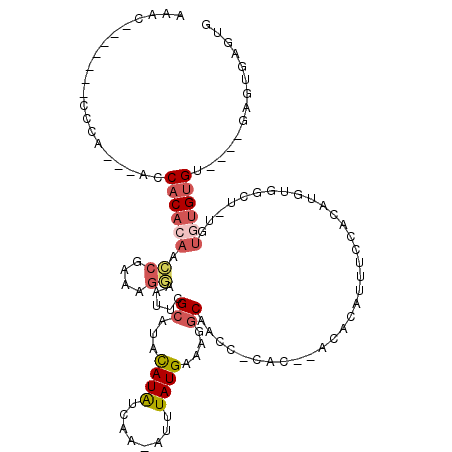
| Location | 19,383,742 – 19,383,851 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.12 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -5.37 |
| Energy contribution | -6.48 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.18 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19383742 109 - 27905053 CACUCACUCUCACACACACACAUAGCCACAUGUGGAAAUGUGUGUGUGAGGUUGCCUUUCAUAAAU-UUGAUAUGUAUGCGAAUUCCUUUCGGUUGUGUGGU---UGGG-------GUUU ..((((..(.(((((((((((((..(((....)))..))))))))))((((((((....((((...-....))))...))))...))))......))).)..---))))-------.... ( -34.30) >DroSec_CAF1 33672 92 - 1 CAC----------ACACACACACAGCCACAUGUGGAAAUGUG-------GGUUGCCUUUCAUAAAU-UUGAUAUGUAUGCGAAUUCCUUUCGGUUGUGUGGU---UGGG-------GUUU ...----------.((..(((((((((((((((.(((.((((-------((......))))))..)-)).))))))....(((.....))))))))))))..---))..-------.... ( -27.60) >DroSim_CAF1 39682 105 - 1 CACUCACUC----ACACACACACAGCCACAUGUGGAAAUGUGUGUGUGAGGUUGCCUUUCAUAAAU-UUGAUAUGUAUGCGAAUUCCUUUCGGUUGUGUGGU---UGGG-------GUUU ......(((----(((((((((...(((....)))...))))))))))))(((((...(((.....-.)))...))).))(((((((..(((......))).---.)))-------)))) ( -34.40) >DroEre_CAF1 38147 103 - 1 CACUCACUC----ACACACACAGAGCCAUACGU------------GUGAGCUUGCCUUUCAUAAAU-UUGACAUGUAUGCGAAUUCCUUUCGCUUGUGUGGUGGUUGGGGUUUUGCCUUU ..((((..(----((.(((((((....((((((------------((.((.((..........)).-)).))))))))(((((.....)))))))))))))))..))))........... ( -31.40) >DroAna_CAF1 41376 85 - 1 AAC----------ACACACACA----------UGAAAGUCGGU--GUG-GGGGGCCCCUCAUAAAU-UUCAUAUGUAUGCGAAUUCCUUUCGGUU-UGUGGU---UGGA-------GUCU ..(----------.((.(((.(----------(((((....((--(.(-((....))).)))...)-))))).))).)).).(((((..(((...-..))).---.)))-------)).. ( -23.30) >DroPer_CAF1 38467 84 - 1 CCCCCAUUC----CCACAU----------------AAAUGUGAGGGAG----GGGGUUCCAUAAAUUUUGAUAUAUAUGCGAAUUUCUCUCCACU--GGGGU---CUGG-------GCUU ..((((..(----((.(((----------------....)))((((((----(..((((((((.((......)).)))).))))..).)))).))--.))).---.)))-------)... ( -28.80) >consensus CACUCACUC____ACACACACA_AGCCACAUGUGGAAAUGUGU__GUG_GGUUGCCUUUCAUAAAU_UUGAUAUGUAUGCGAAUUCCUUUCGGUUGUGUGGU___UGGG_______GUUU ................((((((...............................((....((((........))))...))(((.....)))...)))))).................... ( -5.37 = -6.48 + 1.11)

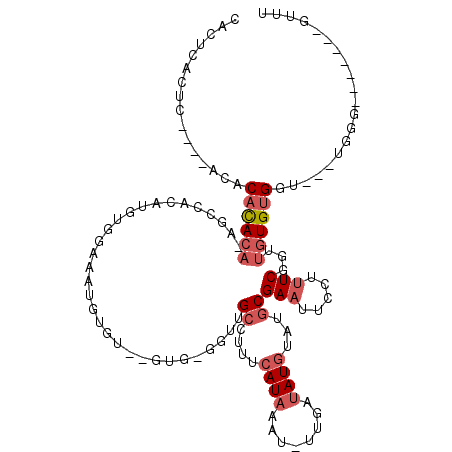
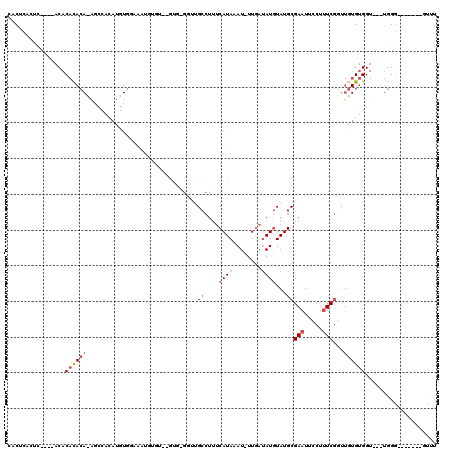
| Location | 19,383,772 – 19,383,876 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -6.87 |
| Energy contribution | -8.05 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.25 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19383772 104 + 27905053 GCAUACAUAUCAAAUUUAUGAAAGGCAACCUCACACACACAUUUCCACAUGUGGCUAUGUGUGUGUGUGAGAGUGAGUGCCCUGGCCAGAACUGAAAUUUAUUU ((...((((.......))))...(((((((((((((((((((..(((....)))....))))))))))))).))...))))...)).................. ( -33.40) >DroSec_CAF1 33702 87 + 1 GCAUACAUAUCAAAUUUAUGAAAGGCAACC-------CACAUUUCCACAUGUGGCUGUGUGUGUGU----------GUGUCCUGGCCAGAGCUGAAAUUUAUUU (((((((((..............(....).-------(((((..(((....)))..))))))))))----------))))...(((....)))........... ( -19.50) >DroSim_CAF1 39712 100 + 1 GCAUACAUAUCAAAUUUAUGAAAGGCAACCUCACACACACAUUUCCACAUGUGGCUGUGUGUGUGU----GAGUGAGUGUCCUGGCCAGAGCUGAAAUUUAUUU ((...((((.......))))..(((((..(((((((((((((..(((....)))..))))))))))----)))....)).))).......))............ ( -33.60) >DroEre_CAF1 38187 88 + 1 GCAUACAUGUCAAAUUUAUGAAAGGCAAGCUCAC------------ACGUAUGGCUCUGUGUGUGU----GAGUGAGUGUCCUGGCCAGAGCUGAAAUUUAUUU ((...((((.......))))..(((((.((((((------------((((((......))))))))----))))...)).))).......))............ ( -22.00) >DroAna_CAF1 41405 81 + 1 GCAUACAUAUGAAAUUUAUGAGGGGCCCCC-CAC--ACCGACUUUCA----------UGUGUGUGU----------GUUUCCUGGCUAUGAAGGAAAUUUAUUU ((((((((((((((.((.((.(((....))-).)--)..)).)))))----------)))))))))----------(((((((........)))))))...... ( -29.90) >consensus GCAUACAUAUCAAAUUUAUGAAAGGCAACCUCAC__ACACAUUUCCACAUGUGGCUGUGUGUGUGU____GAGUGAGUGUCCUGGCCAGAGCUGAAAUUUAUUU ((...((((.......))))..(((....(((((...........(((((......)))))...........)))))...))).)).................. ( -6.87 = -8.05 + 1.18)

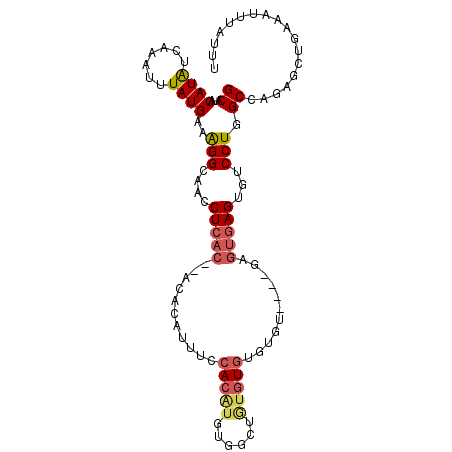
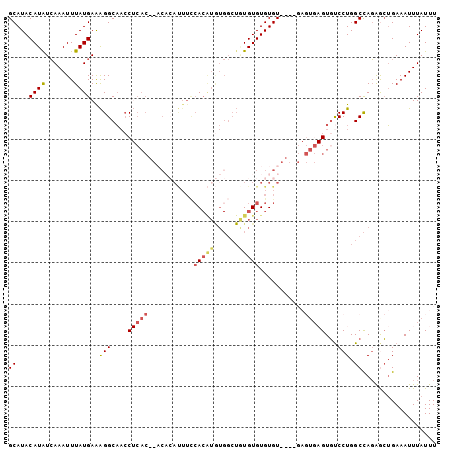
| Location | 19,383,772 – 19,383,876 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -4.18 |
| Energy contribution | -6.23 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.18 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19383772 104 - 27905053 AAAUAAAUUUCAGUUCUGGCCAGGGCACUCACUCUCACACACACACAUAGCCACAUGUGGAAAUGUGUGUGUGAGGUUGCCUUUCAUAAAUUUGAUAUGUAUGC ..(((....((((........((((((...((.((((((((((((.....(((....)))...))))))))))))))))))))........))))....))).. ( -33.59) >DroSec_CAF1 33702 87 - 1 AAAUAAAUUUCAGCUCUGGCCAGGACAC----------ACACACACACAGCCACAUGUGGAAAUGUG-------GGUUGCCUUUCAUAAAUUUGAUAUGUAUGC ...(((((((.......(((...(((.(----------(((....((((......))))....))))-------.))))))......))))))).......... ( -15.72) >DroSim_CAF1 39712 100 - 1 AAAUAAAUUUCAGCUCUGGCCAGGACACUCACUC----ACACACACACAGCCACAUGUGGAAAUGUGUGUGUGAGGUUGCCUUUCAUAAAUUUGAUAUGUAUGC ...(((((((.......(((.((....))..(((----(((((((((...(((....)))...))))))))))))...)))......))))))).......... ( -30.02) >DroEre_CAF1 38187 88 - 1 AAAUAAAUUUCAGCUCUGGCCAGGACACUCACUC----ACACACACAGAGCCAUACGU------------GUGAGCUUGCCUUUCAUAAAUUUGACAUGUAUGC ............((((((.....((.......))----.......))))))(((((((------------((.((.((..........)).)).))))))))). ( -17.90) >DroAna_CAF1 41405 81 - 1 AAAUAAAUUUCCUUCAUAGCCAGGAAAC----------ACACACACA----------UGAAAGUCGGU--GUG-GGGGGCCCCUCAUAAAUUUCAUAUGUAUGC .......((((((........)))))).----------.((.(((.(----------(((((....((--(.(-((....))).)))...)))))).))).)). ( -21.50) >consensus AAAUAAAUUUCAGCUCUGGCCAGGACACUCACUC____ACACACACACAGCCACAUGUGGAAAUGUGU__GUGAGGUUGCCUUUCAUAAAUUUGAUAUGUAUGC ...(((((((.......(((.......(((((...........((((........))))...........)))))...)))......))))))).......... ( -4.18 = -6.23 + 2.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:19 2006