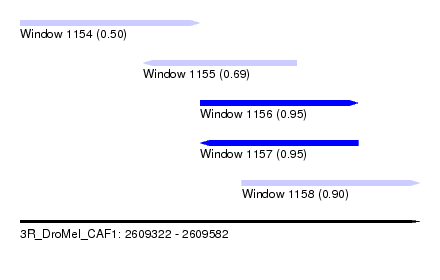
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,609,322 – 2,609,582 |
| Length | 260 |
| Max. P | 0.954972 |

| Location | 2,609,322 – 2,609,439 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -28.35 |
| Energy contribution | -27.97 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2609322 117 + 27905053 UAUAAAGUAUACUAAAGCUUCACGGAAACCUCACAACUAAGCUUCACCUGUCUGUCAACUUGGAGUCUUACAUUUCGGGUGGGUAAAUGGGGAAGUUUAGG-GUU-UGGUG-GGCGGAUG ......((.(((((((.(.....(....).......(((((((((.(((((.(..(.(((((((((.....))))))))).)..).)))))))))))))))-.))-)))))-.))..... ( -34.80) >DroSim_CAF1 73398 119 + 1 UAUAAAGUAUACUAAAGCGGCACGGAAACCUCACAACUAAGCUUCACCUGUCUGUCAACUUGGAGUCUUACAUUUCGGGUGGGGAAAUGGGGAAGUUUUGG-GUUUUGGUGGGGCGGAUG ................((..(((.((((((.((.....(((((((.(((((.(.((.(((((((((.....))))))))).)).).)))))))))))))))-))))).)))..))..... ( -35.00) >DroEre_CAF1 78054 119 + 1 UAUAAAGUAUACGAAAGCGACAUGGAAACCUCACAACUAAGCUUCACCUGUCUGUCAACUUGGAGUCUUACAUUUUGGUUGGGGGAAUGGGAGAGUUUUGG-GUUUUGGUGGGGCGAAUG ................((..(((.((((((.((.....((((((..((..(((.((((((..((........))..)))))).)))..))..)))))))))-))))).)))..))..... ( -34.10) >DroYak_CAF1 76568 109 + 1 -----------CGAAAGCGACACGGAAACCUCGCAACUAAGCUCCACCUGUCUGUCAACUUGGAGUCUUACAUUUUGGGUGGGGAAACGGGGGAGUUUUGGUGUUUUGGUGAAGCGAAUG -----------.....((..(((.(((((...((....(((((((.(((((.(.((.(((..((((.....))))..))).)).).))))))))))))..))))))).)))..))..... ( -37.40) >consensus UAUAAAGUAUACGAAAGCGACACGGAAACCUCACAACUAAGCUUCACCUGUCUGUCAACUUGGAGUCUUACAUUUCGGGUGGGGAAAUGGGGAAGUUUUGG_GUUUUGGUGGGGCGAAUG ................((..(((.(((((.(((.....(((((((.(((((.(.((.(((((((((.....))))))))).)).).))))))))))))))).))))).)))..))..... (-28.35 = -27.97 + -0.38)

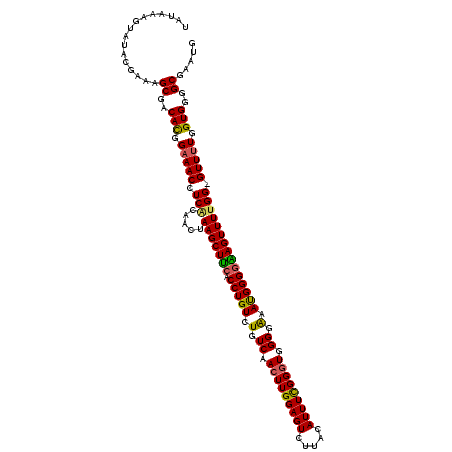
| Location | 2,609,402 – 2,609,502 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2609402 100 - 27905053 GACUGCAUCUGAG----CAGAUGCCUGGCAUAUCUUUUAU-------------GGAGAGCCCGCUCCCCAGACCAUUACUCAUCCGCC-CACCA-AAC-CCUAAACUUCCCCAUUUACCC ....((((((...----.))))))..(((..........(-------------((.(((....))).)))((.......))....)))-.....-...-..................... ( -18.00) >DroSim_CAF1 73478 115 - 1 GACUGCAUCGGAG----CAGAUGCCUGGCAUAUCUUUUAUCUGUCUGGAAACUGGAGAGCCCGCUCCCCAGACCAUCACUCAUCCGCCCCACCAAAAC-CCAAAACUUCCCCAUUUCCCC .........((.(----(.((((((.((((.((.....)).)))).))...((((.(((....))).)))).........)))).)).))........-..................... ( -23.70) >DroEre_CAF1 78134 114 - 1 UACUGCAUCUGAG----CAGAUGCCUGGCAUAUCUUUUAUCUGUCUGGAAACUGGAGAGCCCGCUC-CCAGACCAUCACUCAUUCGCCCCACCAAAAC-CCAAAACUCUCCCAUUCCCCC ....((((((...----.))))))..(((..........(((....)))..((((.(((....)))-))))..............)))..........-..................... ( -22.50) >DroYak_CAF1 76637 119 - 1 UACUGCAUCUGAGAAGCCAGAUGCCUGGCAUGUCUUUUAUCCGUCUGAAAACUGGAGAGCUUGCUC-CCAGACCAUCACUCAUUCGCUUCACCAAAACACCAAAACUCCCCCGUUUCCCC ....((...((((..(((((....)))))......................((((.(((....)))-)))).......))))...))................................. ( -24.10) >consensus GACUGCAUCUGAG____CAGAUGCCUGGCAUAUCUUUUAUCUGUCUGGAAACUGGAGAGCCCGCUC_CCAGACCAUCACUCAUCCGCCCCACCAAAAC_CCAAAACUCCCCCAUUUCCCC ....(((((((......)))))))..((((((.....)))...........((((.(((....))).))))..............)))................................ (-18.05 = -18.42 + 0.38)

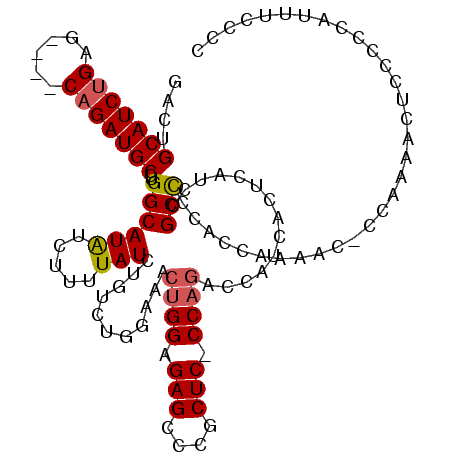
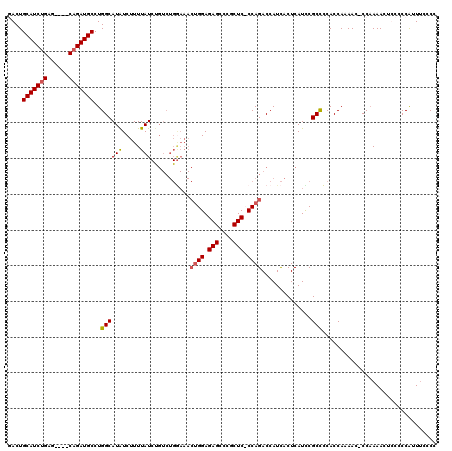
| Location | 2,609,439 – 2,609,542 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -27.85 |
| Energy contribution | -29.10 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2609439 103 + 27905053 AGUAAUGGUCUGGGGAGCGGGCUCUCC-------------AUAAAAGAUAUGCCAGGCAUCUG----CUCAGAUGCAGUCGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAAACGGAAU ..........(((((((....))))))-------------)...........((((((..(((----(......))))..))).(((((....)))))....)))...((....)).... ( -34.40) >DroSim_CAF1 73517 116 + 1 AGUGAUGGUCUGGGGAGCGGGCUCUCCAGUUUCCAGACAGAUAAAAGAUAUGCCAGGCAUCUG----CUCCGAUGCAGUCGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAAACGGAAU .((..(((.((((((((....))))))))...))).))..............((((((..(((----(......))))..))).(((((....)))))....)))...((....)).... ( -42.20) >DroEre_CAF1 78173 109 + 1 AGUGAUGGUCUGG-GAGCGGGCUCUCCAGUUUCCAGACAGAUAAAAGAUAUGCCAGGCAUCUG----CUCAGAUGCAGUAGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAA------A .......((((((-((((.((....)).))))))))))..............((((((..(((----(......))))..))).(((((....)))))....)))........------. ( -38.10) >DroYak_CAF1 76677 113 + 1 AGUGAUGGUCUGG-GAGCAAGCUCUCCAGUUUUCAGACGGAUAAAAGACAUGCCAGGCAUCUGGCUUCUCAGAUGCAGUAGCCAGCAGAUGCAUCUGCGAAAUGGAUAAGGAA------U .......(((((.-.(((..........)))..)))))..........((((((((....)))))(((.((((((((...(....)...)))))))).)))))).........------. ( -34.90) >consensus AGUGAUGGUCUGG_GAGCGGGCUCUCCAGUUUCCAGACAGAUAAAAGAUAUGCCAGGCAUCUG____CUCAGAUGCAGUAGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAA______U .....((((((((((((....))))))))...........................(((((((......)))))))....))))(((((....)))))...................... (-27.85 = -29.10 + 1.25)

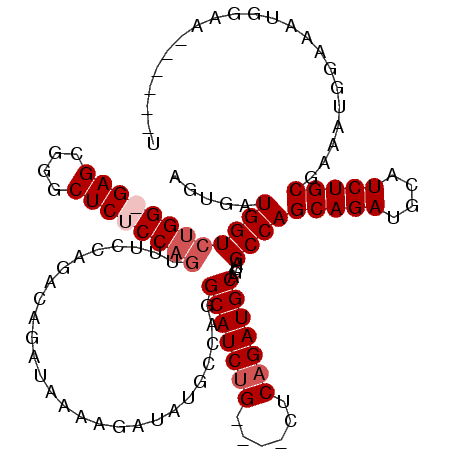
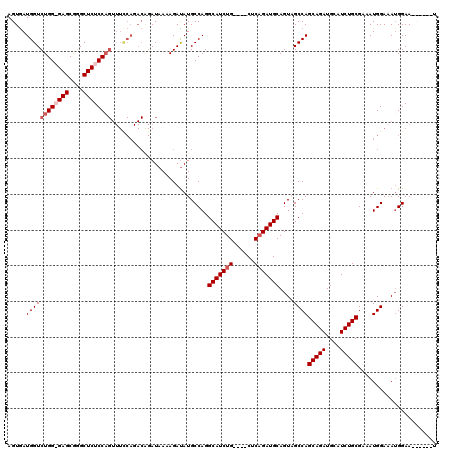
| Location | 2,609,439 – 2,609,542 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.62 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2609439 103 - 27905053 AUUCCGUUUCCAUUUCCAUUUCGCAGAUGCAUCUGCUGGCGACUGCAUCUGAG----CAGAUGCCUGGCAUAUCUUUUAU-------------GGAGAGCCCGCUCCCCAGACCAUUACU ...............(((....(((((....))))).((((.((((......)----))).)))))))...........(-------------((.(((....))).))).......... ( -26.60) >DroSim_CAF1 73517 116 - 1 AUUCCGUUUCCAUUUCCAUUUCGCAGAUGCAUCUGCUGGCGACUGCAUCGGAG----CAGAUGCCUGGCAUAUCUUUUAUCUGUCUGGAAACUGGAGAGCCCGCUCCCCAGACCAUCACU ............((((((....(((((((.((.((((((((.((((......)----))).)))).)))).))....))))))).))))))((((.(((....))).))))......... ( -37.10) >DroEre_CAF1 78173 109 - 1 U------UUCCAUUUCCAUUUCGCAGAUGCAUCUGCUGGCUACUGCAUCUGAG----CAGAUGCCUGGCAUAUCUUUUAUCUGUCUGGAAACUGGAGAGCCCGCUC-CCAGACCAUCACU .------.....((((((....(((((((.((.(((((((..((((......)----)))..))).)))).))....))))))).))))))((((.(((....)))-))))......... ( -35.20) >DroYak_CAF1 76677 113 - 1 A------UUCCUUAUCCAUUUCGCAGAUGCAUCUGCUGGCUACUGCAUCUGAGAAGCCAGAUGCCUGGCAUGUCUUUUAUCCGUCUGAAAACUGGAGAGCUUGCUC-CCAGACCAUCACU .------(((.........(((.((((((((...(....)...)))))))).)))(((((....))))).................)))..((((.(((....)))-))))......... ( -32.70) >consensus A______UUCCAUUUCCAUUUCGCAGAUGCAUCUGCUGGCGACUGCAUCUGAG____CAGAUGCCUGGCAUAUCUUUUAUCUGUCUGGAAACUGGAGAGCCCGCUC_CCAGACCAUCACU ......................(((((....)))))..((((..(((((((......))))))).))))......................((((.(((....))).))))......... (-24.38 = -25.62 + 1.25)

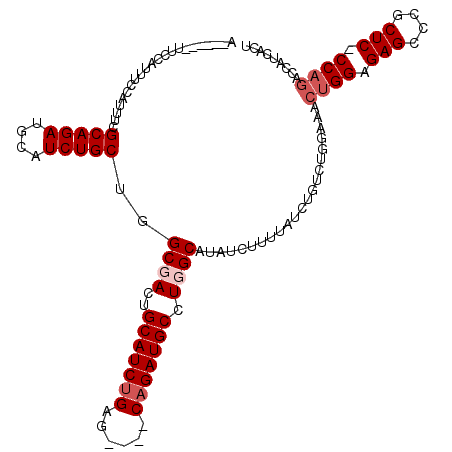
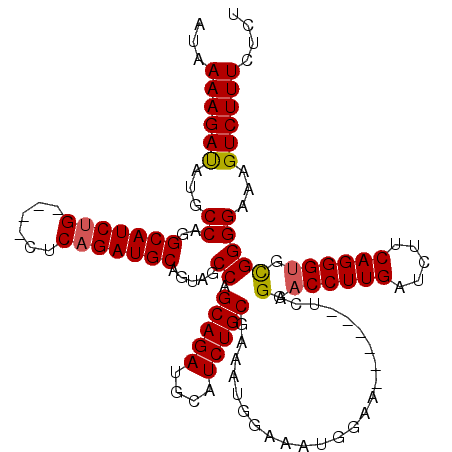
| Location | 2,609,466 – 2,609,582 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -30.55 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2609466 116 + 27905053 AUAAAAGAUAUGCCAGGCAUCUG----CUCAGAUGCAGUCGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAAACGGAAUCGGAACCUUGAUCUUCAGGGCGCGGGGAAAGUCUUUCUCU ...((((((...((..(((((((----((..(((...)))...)))))))))..(((((.....((..((....))...))....(((((.....))))))))))))...)))))).... ( -38.10) >DroSim_CAF1 73557 116 + 1 AUAAAAGAUAUGCCAGGCAUCUG----CUCCGAUGCAGUCGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAAACGGAAUCGGAACCUUGAUCUUCAGGGUGCGGGGAAAGUCUUUCUCU ...((((((...((..(((((((----((.((((...))))..)))))))))..((((......((..((....))...))...((((((.....))))))))))))...)))))).... ( -38.90) >DroEre_CAF1 78212 110 + 1 AUAAAAGAUAUGCCAGGCAUCUG----CUCAGAUGCAGUAGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAA------ACGGAACCUUGAUCUUCAGGGUGCGGGGAAAGUCUUUCUCU ...((((((...((..(((((((----((.....((....)).)))))))))..((((..........((...------.))..((((((.....))))))))))))...)))))).... ( -35.90) >DroYak_CAF1 76716 114 + 1 AUAAAAGACAUGCCAGGCAUCUGGCUUCUCAGAUGCAGUAGCCAGCAGAUGCAUCUGCGAAAUGGAUAAGGAA------UGGGAACCUUGAUCUUCAGGGUGUGGGGAAAGUCUUUCUCU ...((((((...((..((((((((....)))))))).....((((((((....)))))...............------.....((((((.....)))))).)))))...)))))).... ( -35.60) >consensus AUAAAAGAUAUGCCAGGCAUCUG____CUCAGAUGCAGUAGCCAGCAGAUGCAUCUGCGAAAUGGAAAUGGAA______UCGGAACCUUGAUCUUCAGGGUGCGGGGAAAGUCUUUCUCU ...((((((...((..(((((((......))))))).....((.(((((....)))))........................(.((((((.....)))))).)))))...)))))).... (-30.55 = -30.67 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:32 2006