| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,322,134 – 19,322,275 |
| Length | 141 |
| Max. P | 0.999543 |

| Location | 19,322,134 – 19,322,236 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -39.56 |
| Consensus MFE | -26.18 |
| Energy contribution | -28.98 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -5.82 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19322134 102 + 27905053 CAUCCACAUACCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGGCCGAAAUAUGCAACAUGCAUGU---UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCU ..........................(((((((((((((((((((..(((((.(((((((.....))))))---).))))))).))))))))...))))))))). ( -43.60) >DroPse_CAF1 136689 84 + 1 ----AAGA---UC---AGCAGCAGCAGCAACAACAACAACAAGGCCAGGCCAAAAUAUGCAACAUGCAUGUUAUUGUGGC-----------UUGUUGCUGUUGCU ----....---..---...((((((((((((((..((((...(((...)))..(((((((.....))))))).))))...-----------)))))))))))))) ( -34.10) >DroSec_CAF1 117855 102 + 1 CAUCCACAUGCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGCCCAAAAUAUGCAACAUGCAUGU---UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCU ..........................(((((((((((((((((..(((((...(((((((.....))))))---)..)))))..))))))))...))))))))). ( -43.40) >DroSim_CAF1 119212 102 + 1 CAUCCACAUCCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGGCCAAAAUAUGCAACAUGCAUGU---UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCU ..........................(((((((((((((((((((..((((..(((((((.....))))))---)..)))))).))))))))...))))))))). ( -41.00) >DroAna_CAF1 122216 95 + 1 -AACAACA---CC---AGUAACAUUGGUAACAGCAACAACAAGGCCAAGCCAAAAUAUGCAACAUGCAUGU---UAUUGCGGCCGAUGUUGUUGUUGCUGCUGCU -......(---((---(((...))))))..((((((((((((((((..((...(((((((.....))))))---)...))))))....))))))))))))..... ( -35.70) >consensus CAUCCACAU_CCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGGCCAAAAUAUGCAACAUGCAUGU___UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCU ..........................((((((((((((((((((...(((((..((((((.....)))))).....)))))..)))))))))...))))))))). (-26.18 = -28.98 + 2.80)


| Location | 19,322,134 – 19,322,236 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -21.22 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19322134 102 - 27905053 AGCAGCAACAAAAACAACAAGAGCAGCCGCA---ACAUGCAUGUUGCAUAUUUCGGCCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGUAUGUGGAUG .((((((.....(((((((((.((.((((.(---(.((((.....)))).)).))))...)))))))))))..))))))...........(((......)))... ( -36.90) >DroPse_CAF1 136689 84 - 1 AGCAACAGCAACAA-----------GCCACAAUAACAUGCAUGUUGCAUAUUUUGGCCUGGCCUUGUUGUUGUUGUUGCUGCUGCUGCU---GA---UCUU---- ((((.((((((((.-----------...((((((((((((.....)))).....(((...)))..)))))))))))))))).))))...---..---....---- ( -30.30) >DroSec_CAF1 117855 102 - 1 AGCAGCAACAAAAACAACAAGAGCAGCCGCA---ACAUGCAUGUUGCAUAUUUUGGGCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGCAUGUGGAUG .((((((.....(((((((((..(((((.((---(.((((.....))))...))))))))..)))))))))..))))))...........(((......)))... ( -36.90) >DroSim_CAF1 119212 102 - 1 AGCAGCAACAAAAACAACAAGAGCAGCCGCA---ACAUGCAUGUUGCAUAUUUUGGCCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGGAUGUGGAUG .((((((.....(((((((((.((.((((.(---(.((((.....)))).)).))))...)))))))))))..))))))...........(((......)))... ( -35.30) >DroAna_CAF1 122216 95 - 1 AGCAGCAGCAACAACAACAUCGGCCGCAAUA---ACAUGCAUGUUGCAUAUUUUGGCUUGGCCUUGUUGUUGCUGUUACCAAUGUUACU---GG---UGUUGUU- (((((((((((((((((....((((((...(---(.((((.....)))).))...))..))))))))))))))))))((((.......)---))---))))...- ( -35.20) >consensus AGCAGCAACAAAAACAACAAGAGCAGCCGCA___ACAUGCAUGUUGCAUAUUUUGGCCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGG_AUGUGGAUG (((((((((((.........................((((.....)))).....(((...)))...)))))))))))............................ (-21.22 = -20.62 + -0.60)

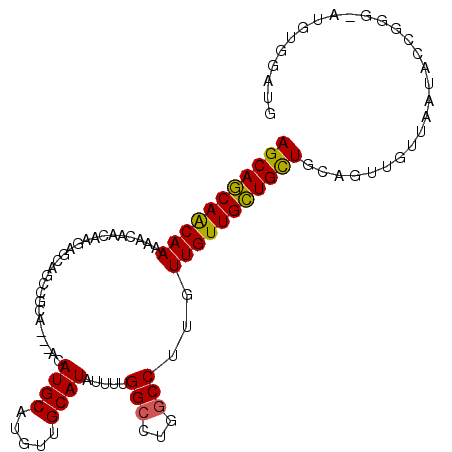
| Location | 19,322,171 – 19,322,275 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.36 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19322171 104 + 27905053 AACAAGGCCA-GGCCGAAAUAUGCAACAUGCAUGU---UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUAGCCACUGCAAACCCAAGGGGGGAAACUGCAUUC ((((((((..-(((((.(((((((.....))))))---).))))))).))))))......((.(((((....))))).))(((...((....))((....)))))... ( -39.40) >DroVir_CAF1 133708 88 + 1 AACAAGGCCGCAACAAAAAUAUGCAGCAUGCAUGUUGUU---UUUGUAACUGUUGUUGUUACUGAUGUUGUGGUCGCCAAAGCAAAGCAAA----------------- .....((((((((((..(((((((.....)))))))...---...(((((.......)))))...))))))))))((....))........----------------- ( -27.70) >DroSec_CAF1 117892 104 + 1 AACAAGGCCA-GCCCAAAAUAUGCAACAUGCAUGU---UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUUGCCACUGCAAACCCAAGGGGGGAAACUGCAUCC ((((((..((-(((...(((((((.....))))))---)..)))))..))))))........((((((.((((...)))).)))..((....))((....)))))... ( -38.30) >DroSim_CAF1 119249 104 + 1 AACAAGGCCA-GGCCAAAAUAUGCAACAUGCAUGU---UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUUGCCACUGCAAACCCAAGGGGGGAAACUGCAUCC ((((((((..-((((..(((((((.....))))))---)..)))))).))))))........((((((.((((...)))).)))..((....))((....)))))... ( -35.90) >DroAna_CAF1 122246 104 + 1 AACAAGGCCA-AGCCAAAAUAUGCAACAUGCAUGU---UAUUGCGGCCGAUGUUGUUGUUGCUGCUGCUGCGGUCGCCAAAGCCAACCGAAGGGGGGAAAAUGCAUUC ((((.((((.-.((...(((((((.....))))))---)...))))))..))))......(((((....))))).((.....((..((....)).)).....)).... ( -30.60) >consensus AACAAGGCCA_GGCCAAAAUAUGCAACAUGCAUGU___UGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUCGCCACUGCAAACCCAAGGGGGGAAACUGCAUCC .....(((((.(((...((((.((((((.(((............)))...))))))...))))...))).)))))((....))...((....)).............. (-16.00 = -16.36 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:57 2006