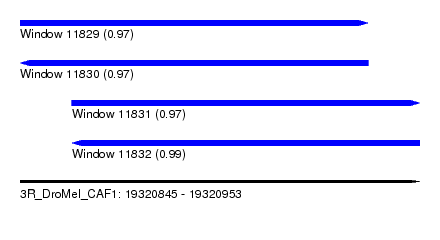
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,320,845 – 19,320,953 |
| Length | 108 |
| Max. P | 0.994610 |

| Location | 19,320,845 – 19,320,939 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -21.93 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19320845 94 + 27905053 --------------------------CAAUGGCAGACAGUUUCCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAA --------------------------...(((.(((...))))))....(((((((((((.((.((((((((.....)))))))).)).................))))))))))).... ( -24.17) >DroPse_CAF1 135326 120 + 1 GUACAGGGGACAGCUAACAGAGGACAUAUAGGCAGACAGUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAA .....((..((..((..(............)..))...))..)).....(((((((((((.(((((((((((.....)))))))).)))................))))))))))).... ( -31.23) >DroWil_CAF1 167550 84 + 1 ------------------------------------CAAUUUCCAACUAAUUAGGCUUGCUGGCUUUUAAUCAAAACGAUUAAAGUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAA ------------------------------------.............(((((((..(..(((((((((((.....)))))))).)))..)(((.........)))..))))))).... ( -18.80) >DroMoj_CAF1 133503 86 + 1 ----------------------------------GAAAACUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAA ----------------------------------...............(((((((((((.(((((((((((.....)))))))).)))................))))))))))).... ( -24.73) >DroAna_CAF1 120925 91 + 1 -----------------------------AGGCACACAAUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAA -----------------------------....................(((((((((((.(((((((((((.....)))))))).)))................))))))))))).... ( -24.73) >DroPer_CAF1 135555 120 + 1 GGACAGGGGACAGGGGACAGAGGACAUAUAGGCAGACAGUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAA .....((..((.(....)............(.....).))..)).....(((((((((((.(((((((((((.....)))))))).)))................))))))))))).... ( -33.33) >consensus _____________________________AGGCAGACAAUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAA .................................................(((((((((((.(((((((((((.....)))))))).)))................))))))))))).... (-21.93 = -22.30 + 0.36)

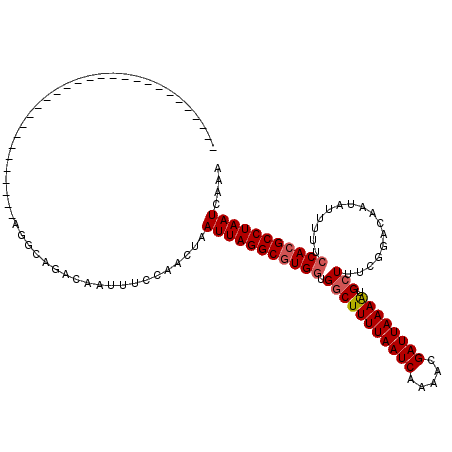

| Location | 19,320,845 – 19,320,939 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19320845 94 - 27905053 UUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGGAAACUGUCUGCCAUUG-------------------------- .((((((((((((((.......((....))..((.((((((((.....)))))))).)).))))))))))))))..(....)............-------------------------- ( -28.90) >DroPse_CAF1 135326 120 - 1 UUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCCUAUAUGUCCUCUGUUAGCUGUCCCCUGUAC .((((((((((((((............(....)..((((((((.....))))))))....))))))))))))))..((..((.((..((.............))..)).))..))..... ( -29.52) >DroWil_CAF1 167550 84 - 1 UUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCACUUUAAUCGUUUUGAUUAAAAGCCAGCAAGCCUAAUUAGUUGGAAAUUG------------------------------------ .((((((((((.(((.........((.(....).))(((((((.....)))))))..)))....))))))))))..........------------------------------------ ( -19.80) >DroMoj_CAF1 133503 86 - 1 UUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAGUUUUC---------------------------------- .((((((((((((((........(((......)))((((((((.....))))))))....))))))))))))))............---------------------------------- ( -26.50) >DroAna_CAF1 120925 91 - 1 UUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAAUUGUGUGCCU----------------------------- .((((((((((((((............(....)..((((((((.....))))))))....)))))))))))))).................----------------------------- ( -27.10) >DroPer_CAF1 135555 120 - 1 UUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCCUAUAUGUCCUCUGUCCCCUGUCCCCUGUCC .((((((((((((((............(....)..((((((((.....))))))))....))))))))))))))..(((...(((......)))..)))..................... ( -28.40) >consensus UUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCCU_____________________________ .((((((((((((((........(((......)))((((((((.....))))))))....)))))))))))))).............................................. (-23.98 = -24.48 + 0.50)



| Location | 19,320,859 – 19,320,953 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.56 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -19.78 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19320859 94 + 27905053 UUCCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU-------------------------- .........(((((((((((.((.((((((((.....)))))))).)).................)))))))))))..................-------------------------- ( -23.87) >DroVir_CAF1 132531 94 + 1 UUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU-------------------------- .........(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................-------------------------- ( -24.73) >DroGri_CAF1 126240 94 + 1 UUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCGACGCCUAAUCAAAGACUUAAUACCAUU-------------------------- .........(((((((((((.(((((((((((.....)))))))).)))..))((.........)).)))))))))..................-------------------------- ( -21.90) >DroSim_CAF1 117945 94 + 1 UUCCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU-------------------------- .........(((((((((((.((.((((((((.....)))))))).)).................)))))))))))..................-------------------------- ( -23.87) >DroWil_CAF1 167554 120 + 1 UUCCAACUAAUUAGGCUUGCUGGCUUUUAAUCAAAACGAUUAAAGUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGAGUUUCGGUUUUUUCGAUGUGGUUGUUGUUGUAGUUUUA ...(((((((((((((..(..(((((((((((.....)))))))).)))..)(((.........)))..))))))).........((((.....))))..)))))).............. ( -26.00) >DroMoj_CAF1 133509 94 + 1 UUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU-------------------------- .........(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................-------------------------- ( -24.73) >consensus UUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU__________________________ .........(((((((((((.(((((((((((.....)))))))).)))................)))))))))))............................................ (-19.78 = -20.48 + 0.70)


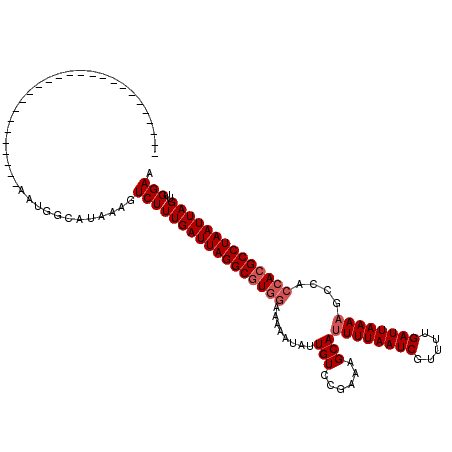
| Location | 19,320,859 – 19,320,953 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.56 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -23.80 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19320859 94 - 27905053 --------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGGAA --------------------------...(((.....))).((((((((((((((.......((....))..((.((((((((.....)))))))).)).))))))))))))))...... ( -29.00) >DroVir_CAF1 132531 94 - 1 --------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAA --------------------------...(((.....))).((((((((((((((........(((......)))((((((((.....))))))))....))))))))))))))...... ( -27.60) >DroGri_CAF1 126240 94 - 1 --------------------------AAUGGUAUUAAGUCUUUGAUUAGGCGUCGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAA --------------------------............((((((((((((((((((......)))..((...((.((((((((.....))))))))))..))))))))))))))..))). ( -23.00) >DroSim_CAF1 117945 94 - 1 --------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGGAA --------------------------...(((.....))).((((((((((((((.......((....))..((.((((((((.....)))))))).)).))))))))))))))...... ( -29.00) >DroWil_CAF1 167554 120 - 1 UAAAACUACAACAACAACCACAUCGAAAAAACCGAAACUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCACUUUAAUCGUUUUGAUUAAAAGCCAGCAAGCCUAAUUAGUUGGAA ...........((((.......(((.......))).......(((((((((.(((.........((.(....).))(((((((.....)))))))..)))....)))))))))))))... ( -24.70) >DroMoj_CAF1 133509 94 - 1 --------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAA --------------------------...(((.....))).((((((((((((((........(((......)))((((((((.....))))))))....))))))))))))))...... ( -27.60) >consensus __________________________AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAA ......................................(((((((((((((((((........(((......)))((((((((.....))))))))....))))))))))))))..))). (-23.80 = -24.47 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:54 2006