
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,307,742 – 19,307,897 |
| Length | 155 |
| Max. P | 0.943099 |

| Location | 19,307,742 – 19,307,858 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.29 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19307742 116 + 27905053 UCUGAUAACUCAACGGCUUUAUUGUGGAAUGAUCAGACAUACCCUCACUAUCUAAGACGCCGGCAGGAUACCGACAUAUUGGUAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGA ((((((...((.((((.....)))).))...)))))).....(((((((.....)).....((((((((((..((......)).))))))))))))))).....((.......)). ( -31.60) >DroSec_CAF1 103553 116 + 1 UCUGAUAACUCAACGGCUUUAUUGUGGAAUGAUCAAACAUACCCACAUUAUCUCAGACACCGGCAGGAUACCGACAUAUUGGCAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGA ..(((...((((..((...((.(((((.(((......)))..))))).))..)).......((((((((((.(.(.....).).))))))))))))))...)))((.......)). ( -30.90) >DroSim_CAF1 106515 116 + 1 UCUGAUAACUCAACGGCUUUAUUGUGGAAUGAUCAAACAUACCCACAUUAUCUGAGACACCGGCAGGAUACCGACAUAUUGGCAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGA ((((...............((.(((((.(((......)))..))))).))..((((((..(((((((((((.(.(.....).).)))))))))).)..)))))).......)))). ( -33.70) >DroEre_CAF1 106841 116 + 1 UCUGCCGACUCAACGGCUUUAAUGUGGAAUAAUCAAACGUACCCACAUUAUCUGAGGCAGCGACAGGAUUCCGACAUGUUGUCAGUGUCCUGCCUGAGAUUUCACCAGUACCAGGA ...((((......))))..((((((((...............))))))))(((.((((((.((((((...))(((.....)))..)))))))))).))).....((.......)). ( -37.06) >DroYak_CAF1 108995 116 + 1 ACUGACAACUCAACUGCUUUAAUGUGGAAUAAUCAGACAUAUCCACAUUAUCUGCGACACCGACAGGAUGCCGACAUAUUGGCAGUGUCCUGUCUGAGAUUUCAGCAAUAUCAGGA .((((...((((..(((..(((((((((.............)))))))))...))).....(((((((((((((....))))))...)))))))))))...))))........... ( -37.52) >DroAna_CAF1 105476 116 + 1 AGUAGCAAUGUCACCACACUGGUCUGGGAUGAUCAGGAGUAUCCACAUUACCUACGGCACCACCAAGACGUGGAUCUAAUUGGAAUGGUAUGUCUAAAGACUUCUCAAUAUGGAGG .((((.(((((....((.((((((......))))))..))....)))))..))))((((((((......)))).(((....)))......))))......((((.......)))). ( -25.40) >consensus UCUGACAACUCAACGGCUUUAUUGUGGAAUGAUCAAACAUACCCACAUUAUCUGAGACACCGGCAGGAUACCGACAUAUUGGCAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGA ..(((...((((..........(((((...............)))))..............((((((((((...(.....)...))))))))))))))...)))............ (-17.95 = -18.29 + 0.34)

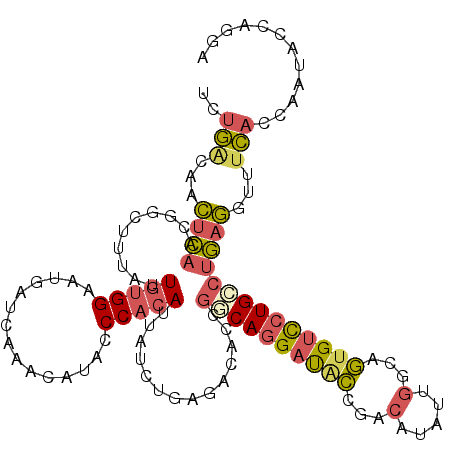

| Location | 19,307,742 – 19,307,858 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.89 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19307742 116 - 27905053 UCCUGGUAUUGGUGAAACCUCAGGCAGGACACUACCAAUAUGUCGGUAUCCUGCCGGCGUCUUAGAUAGUGAGGGUAUGUCUGAUCAUUCCACAAUAAAGCCGUUGAGUUAUCAGA ........(((((((...(((((((((((...((((........)))))))))))(((..((((.....))))((.(((......))).))........)))..))))))))))). ( -33.90) >DroSec_CAF1 103553 116 - 1 UCCUGGUAUUGGUGAAACCUCAGGCAGGACACUGCCAAUAUGUCGGUAUCCUGCCGGUGUCUGAGAUAAUGUGGGUAUGUUUGAUCAUUCCACAAUAAAGCCGUUGAGUUAUCAGA ..((((((.((((((.(((...(((((((.((((.(.....).)))).)))))))))).))........((((((.(((......))))))))).....))))......)))))). ( -37.40) >DroSim_CAF1 106515 116 - 1 UCCUGGUAUUGGUGAAACCUCAGGCAGGACACUGCCAAUAUGUCGGUAUCCUGCCGGUGUCUCAGAUAAUGUGGGUAUGUUUGAUCAUUCCACAAUAAAGCCGUUGAGUUAUCAGA ..((((((.((((((.(((...(((((((.((((.(.....).)))).)))))))))).))........((((((.(((......))))))))).....))))......)))))). ( -37.40) >DroEre_CAF1 106841 116 - 1 UCCUGGUACUGGUGAAAUCUCAGGCAGGACACUGACAACAUGUCGGAAUCCUGUCGCUGCCUCAGAUAAUGUGGGUACGUUUGAUUAUUCCACAUUAAAGCCGUUGAGUCGGCAGA .((.......)).....(((.(((((((((((((((.....))))).....)))).)))))).)))(((((((((.............)))))))))..((((......))))... ( -38.82) >DroYak_CAF1 108995 116 - 1 UCCUGAUAUUGCUGAAAUCUCAGACAGGACACUGCCAAUAUGUCGGCAUCCUGUCGGUGUCGCAGAUAAUGUGGAUAUGUCUGAUUAUUCCACAUUAAAGCAGUUGAGUUGUCAGU ..........(((((...(((((((((((...((((........)))))))))))......((...(((((((((.(((......))))))))))))..))...))))...))))) ( -40.00) >DroAna_CAF1 105476 116 - 1 CCUCCAUAUUGAGAAGUCUUUAGACAUACCAUUCCAAUUAGAUCCACGUCUUGGUGGUGCCGUAGGUAAUGUGGAUACUCCUGAUCAUCCCAGACCAGUGUGGUGACAUUGCUACU .(.((((((((....(((....)))......................((((.((.((((...((((....((....)).))))..)))))))))))))))))).)........... ( -26.30) >consensus UCCUGGUAUUGGUGAAACCUCAGGCAGGACACUGCCAAUAUGUCGGUAUCCUGCCGGUGUCUCAGAUAAUGUGGGUAUGUCUGAUCAUUCCACAAUAAAGCCGUUGAGUUAUCAGA ......................(((((((...((((........)))))))))))((((.(((((....((((((.(((......))))))))).........))))).))))... (-17.88 = -18.89 + 1.01)


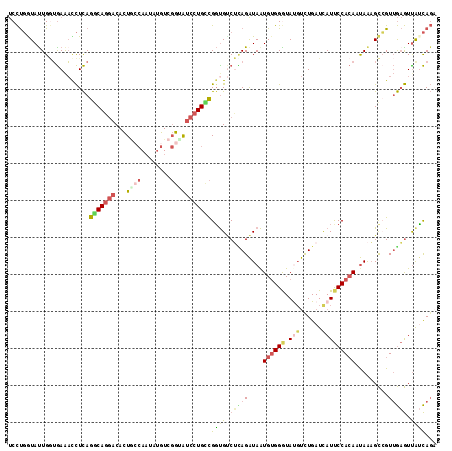
| Location | 19,307,780 – 19,307,897 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19307780 117 + 27905053 AUACCCUCACUAUCUAAGACGCCGGCAGGAUACCGACAUAUUGGUAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGAAGUACUCUUAGCAUUAAGUUUAAUGUUGGAUCAAAUGC-G .((((((((((.....)).....((((((((((..((......)).))))))))))))))).....((.......))..)))......(((((..(((((....)))))..)))))-. ( -30.10) >DroSec_CAF1 103591 117 + 1 AUACCCACAUUAUCUCAGACACCGGCAGGAUACCGACAUAUUGGCAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGAAGUACUCCUAGCAUUAAGCUUAAUGUUGGAUCAAAUGC-G .......((((......((.(((((((((((((.(.(.....).).))))))))))...))).)).((((((..((((.....))))(((.....)))...))))))....)))).-. ( -32.80) >DroSim_CAF1 106553 118 + 1 AUACCCACAUUAUCUGAGACACCGGCAGGAUACCGACAUAUUGGCAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGAAGUACUCCUAGCAUUAAGCUUAAUGUUGGAUCAAAUGCCC .......((((...((((((..(((((((((((.(.(.....).).)))))))))).)..))))))((((((..((((.....))))(((.....)))...))))))....))))... ( -36.20) >DroEre_CAF1 106879 117 + 1 GUACCCACAUUAUCUGAGGCAGCGACAGGAUUCCGACAUGUUGUCAGUGUCCUGCCUGAGAUUUCACCAGUACCAGGAAGUGCUGCUAGCAUUAAGCGUAAUGCUGGAUCAAAUAC-U ...........((((.((((((.((((((...))(((.....)))..)))))))))).)))).....((((((......))))))(((((((((....))))))))).........-. ( -39.40) >DroYak_CAF1 109033 117 + 1 AUAUCCACAUUAUCUGCGACACCGACAGGAUGCCGACAUAUUGGCAGUGUCCUGUCUGAGAUUUCAGCAAUAUCAGGAAGUACUGCUAGCAUUAAGCGCAAUGCUGGAUCAAAUAC-G ...(((....(((.(((......(((((((((((((....))))))...)))))))((......))))))))...))).(((.(.((((((((......))))))))....).)))-. ( -32.40) >DroAna_CAF1 105514 117 + 1 GUAUCCACAUUACCUACGGCACCACCAAGACGUGGAUCUAAUUGGAAUGGUAUGUCUAAAGACUUCUCAAUAUGGAGGGGUAAUGAACGCCCUCAAGGUAAUGUUGAAUCAAAUGA-G ...((.(((((((((..(((.((((......))))......(((((........)))))..(((((((......))))))).......)))....))))))))).)).........-. ( -30.90) >consensus AUACCCACAUUAUCUGAGACACCGGCAGGAUACCGACAUAUUGGCAGUGUCCUGCCUGAGGUUUCACCAAUACCAGGAAGUACUCCUAGCAUUAAGCGUAAUGUUGGAUCAAAUGC_G ............(((........((((((((((...(.....)...))))))))))...(((.........))).))).......(((((((((....)))))))))........... (-17.54 = -18.47 + 0.92)


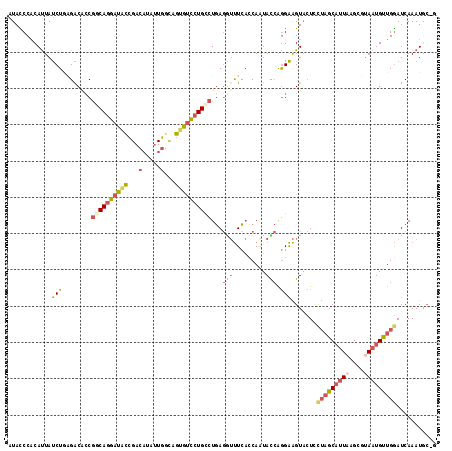
| Location | 19,307,780 – 19,307,897 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19307780 117 - 27905053 C-GCAUUUGAUCCAACAUUAAACUUAAUGCUAAGAGUACUUCCUGGUAUUGGUGAAACCUCAGGCAGGACACUACCAAUAUGUCGGUAUCCUGCCGGCGUCUUAGAUAGUGAGGGUAU (-((.((((((.....))))))..((((((((.(((...))).)))))))))))...((((((((((((...((((........)))))))))))...(((...)))..))))).... ( -31.10) >DroSec_CAF1 103591 117 - 1 C-GCAUUUGAUCCAACAUUAAGCUUAAUGCUAGGAGUACUUCCUGGUAUUGGUGAAACCUCAGGCAGGACACUGCCAAUAUGUCGGUAUCCUGCCGGUGUCUGAGAUAAUGUGGGUAU .-.......(((((.(((((.(((.(((((((((((...))))))))))))))((.(((...(((((((.((((.(.....).)))).)))))))))).)).....)))))))))).. ( -42.40) >DroSim_CAF1 106553 118 - 1 GGGCAUUUGAUCCAACAUUAAGCUUAAUGCUAGGAGUACUUCCUGGUAUUGGUGAAACCUCAGGCAGGACACUGCCAAUAUGUCGGUAUCCUGCCGGUGUCUCAGAUAAUGUGGGUAU .........(((((.(((((....((((((((((((...))))))))))))(.((.(((...(((((((.((((.(.....).)))).)))))))))).)).)...)))))))))).. ( -42.40) >DroEre_CAF1 106879 117 - 1 A-GUAUUUGAUCCAGCAUUACGCUUAAUGCUAGCAGCACUUCCUGGUACUGGUGAAAUCUCAGGCAGGACACUGACAACAUGUCGGAAUCCUGUCGCUGCCUCAGAUAAUGUGGGUAC .-.(((((((...(((((((....))))))).(((((..(((((......)).)))......(((((((..(((((.....)))))..)))))))))))).))))))).......... ( -39.10) >DroYak_CAF1 109033 117 - 1 C-GUAUUUGAUCCAGCAUUGCGCUUAAUGCUAGCAGUACUUCCUGAUAUUGCUGAAAUCUCAGACAGGACACUGCCAAUAUGUCGGCAUCCUGUCGGUGUCGCAGAUAAUGUGGAUAU .-.......(((((.((((((((...(((((((((((((.....).))))))).........(((((((...((((........)))))))))))))))).)).).)))))))))).. ( -34.90) >DroAna_CAF1 105514 117 - 1 C-UCAUUUGAUUCAACAUUACCUUGAGGGCGUUCAUUACCCCUCCAUAUUGAGAAGUCUUUAGACAUACCAUUCCAAUUAGAUCCACGUCUUGGUGGUGCCGUAGGUAAUGUGGAUAC .-.......(((((.((((((((.(((((.((.....)))))))...........(((....))).(((((..(((...((((....))))))))))))....))))))))))))).. ( -31.40) >consensus C_GCAUUUGAUCCAACAUUAAGCUUAAUGCUAGCAGUACUUCCUGGUAUUGGUGAAACCUCAGGCAGGACACUGCCAAUAUGUCGGUAUCCUGCCGGUGUCUCAGAUAAUGUGGGUAU .........((((.((((((.(((.((((((((.........))))))))))).........(((((((...((((........)))))))))))...........)))))))))).. (-22.04 = -22.52 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:51 2006