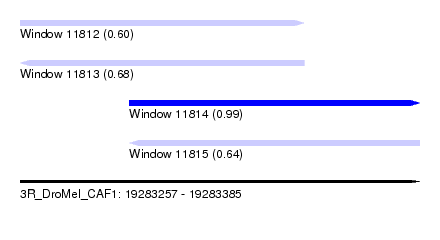
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,283,257 – 19,283,385 |
| Length | 128 |
| Max. P | 0.994747 |

| Location | 19,283,257 – 19,283,348 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -17.80 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19283257 91 + 27905053 CGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCA----AAUGGCCGUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGU ((((((....(..(.....)..)....)))))).......----..((((((.(((.((((((((......)))))))).....))))))))--------------).. ( -29.70) >DroPse_CAF1 94436 90 + 1 ----CAAUUUGUGGUUUCUUUGUUUCCUGACCGAGUCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAGUC-AAAUGACUGCG--------------AGG ----...((((..(((..((((....(((.((((((.......((((((((((....).))))))))))))))).))).)-))).)))..))--------------)). ( -28.21) >DroEre_CAF1 80383 91 + 1 CGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCA----AAUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGC (((((((((.((((((..((((...((.....).)...))----)).))))))...))))))))).....((((((.((......)).))))--------------)). ( -26.10) >DroYak_CAF1 83676 91 + 1 CGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCA----AAUGGCCAUGCUAGUUGGCCACUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGG .........................((((((((.((....----..((((...))))((((((((......))))))))......)))))))--------------))) ( -28.40) >DroAna_CAF1 80482 108 + 1 GGGUCAAUUUGUGGUUUCUUUGUUUCCUGGCUGCGUCCCA-UAUAAUGGCCACGUUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGGCGGCCUGUGUGUGGCUGCGAGG .(((((....(..(.....)..)....)))))(((..(((-((((..((((.(.(((((((((((......))))))))...))).).))))..))))))).))).... ( -37.30) >DroPer_CAF1 94542 90 + 1 ----CAAUUUGUGGUUUCUUUGUUUCCUGACCGAGUCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAGUC-AAAUGACUGCG--------------AGG ----...((((..(((..((((....(((.((((((.......((((((((((....).))))))))))))))).))).)-))).)))..))--------------)). ( -28.21) >consensus CGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCA____AAUGGCCACGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGCC______________AGG ..........((((((............(((...)))..........))))))....((((((((......)))))))).............................. (-17.80 = -16.92 + -0.89)

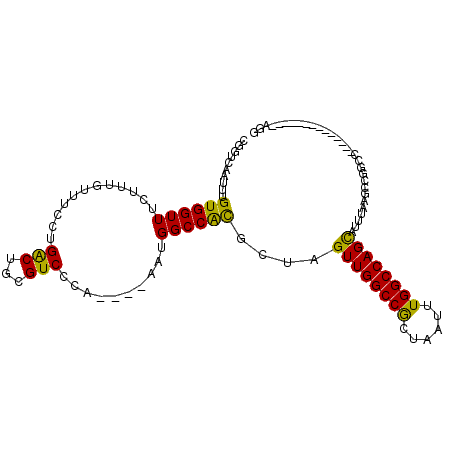

| Location | 19,283,257 – 19,283,348 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19283257 91 - 27905053 ACU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCACGGCCAUU----UGGGACGCAGUCAGGAAACAAAGAAACCACAAAUUGACCG .((--------------(((((((......)))).((((((....((((.........)))))))----)))......))))).......................... ( -22.90) >DroPse_CAF1 94436 90 - 1 CCU--------------CGCAGUCAUUU-GACUGGCCAAAUUAACGGCCAACUAACGUGGCCAUUAUAUGGGGACUCGGUCAGGAAACAAAGAAACCACAAAUUG---- (((--------------(((((((....-)))))...........(((((.(....))))))......)))))....(((..(....)......)))........---- ( -24.20) >DroEre_CAF1 80383 91 - 1 GCU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCAUGGCCAUU----UGGGACGCAGUCAGGAAACAAAGAAACCACAAAUUGACCG (((--------------(((((((......))))).....)))))(((((.......)))))...----((((((...))).(....).......)))........... ( -25.40) >DroYak_CAF1 83676 91 - 1 CCU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGUGGCCAACUAGCAUGGCCAUU----UGGGACGCAGUCAGGAAACAAAGAAACCACAAAUUGACCG (((--------------(((((((......)))).((((....(((((((.......))))))))----)))......))))))......................... ( -27.90) >DroAna_CAF1 80482 108 - 1 CCUCGCAGCCACACACAGGCCGCCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAACGUGGCCAUUAUA-UGGGACGCAGCCAGGAAACAAAGAAACCACAAAUUGACCC (((.((.(((.......)))(((((...(((.((((((..((((.......))))..)))))).))).-.))).))..)).)))......................... ( -27.30) >DroPer_CAF1 94542 90 - 1 CCU--------------CGCAGUCAUUU-GACUGGCCAAAUUAACGGCCAACUAACGUGGCCAUUAUAUGGGGACUCGGUCAGGAAACAAAGAAACCACAAAUUG---- (((--------------(((((((....-)))))...........(((((.(....))))))......)))))....(((..(....)......)))........---- ( -24.20) >consensus CCU______________GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAACAUGGCCAUU____UGGGACGCAGUCAGGAAACAAAGAAACCACAAAUUGACCG .................(((((.((((.....((((((..((((.......))))..))))))......)))).))..))).(....)..................... (-15.23 = -14.87 + -0.36)

| Location | 19,283,292 – 19,283,385 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -17.04 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19283292 93 + 27905053 UCCCA----AAUGGCCGUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAAC-AAAGGCCACACUCUAAUUGAUAAAUU ...((----(.((((((.(((.((((((((......)))))))).....))))))))--------------)((((((.......-))))))..........)))....... ( -27.80) >DroVir_CAF1 98621 96 + 1 UCCCCA--UAACGGCCACCUUGGUUGGCCACUAAUUUGGCCGGCAAUAAGACCACGC----------UAC-AUGGCUUAAUUAAA---AAGUAACUCUCUAAUUGAUUAAUU ...(((--(...(((...((((((((((((......))))))))..)))).....))----------)..-)))).((((((((.---.((.......))..)))))))).. ( -20.20) >DroPse_CAF1 94467 97 + 1 UCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAGUC-AAAUGACUGCG--------------AGGCUUUAAUUAAAAAAAGGCCACACUCUAAUUGAUAAAUU ...(((..((((((((((....).)))))))))...)))((((((-....))))).)--------------.((((((.........))))))................... ( -27.30) >DroYak_CAF1 83711 93 + 1 UCCCA----AAUGGCCAUGCUAGUUGGCCACUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGGCUUUAAUUAAC-AAAGGCCACACUCUAAUUGAUAAAUU .....----..(((((......((((((((......))))))))(((.(((((....--------------.))))).)))....-...))))).................. ( -30.10) >DroAna_CAF1 80517 110 + 1 UCCCA-UAUAAUGGCCACGUUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGGCGGCCUGUGUGUGGCUGCGAGGCUUUAAUUAAC-AAAGGCCACACUCUAAUUGAUAAAUU ..(((-((((..((((.(.(((((((((((......))))))))...))).).))))..)))))))......((((((.......-.))))))................... ( -37.50) >DroPer_CAF1 94573 97 + 1 UCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAGUC-AAAUGACUGCG--------------AGGCUUUAAUUAAAAAAAGGCCACACUCUAAUUGAUAAAUU ...(((..((((((((((....).)))))))))...)))((((((-....))))).)--------------.((((((.........))))))................... ( -27.30) >consensus UCCCA___UAAUGGCCACGUUAGUUGGCCGCUAAUUUGGCCAGCAUUAAAGCCGGCC______________AGGCUUUAAUUAAA_AAAGGCCACACUCUAAUUGAUAAAUU ...........(((((......((((((((......))))))))..........((.................))..............))))).................. (-17.04 = -16.80 + -0.25)



| Location | 19,283,292 – 19,283,385 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -13.28 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19283292 93 - 27905053 AAUUUAUCAAUUAGAGUGUGGCCUUU-GUUAAUUAAAGACU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCACGGCCAUU----UGGGA .................((((((..(-(((((((......(--------------(.(((((......))))).))))))))))((......))..)))))).----..... ( -23.20) >DroVir_CAF1 98621 96 - 1 AAUUAAUCAAUUAGAGAGUUACUU---UUUAAUUAAGCCAU-GUA----------GCGUGGUCUUAUUGCCGGCCAAAUUAGUGGCCAACCAAGGUGGCCGUUA--UGGGGA ........((((((((((...)))---)))))))...((..-(((----------((.(((((........))))).......(((((.(....))))))))))--)..)). ( -26.60) >DroPse_CAF1 94467 97 - 1 AAUUUAUCAAUUAGAGUGUGGCCUUUUUUUAAUUAAAGCCU--------------CGCAGUCAUUU-GACUGGCCAAAUUAACGGCCAACUAACGUGGCCAUUAUAUGGGGA ...............(((.(((.(((........)))))).--------------)))((((....-))))..(((...(((.(((((.(....)))))).)))..)))... ( -24.80) >DroYak_CAF1 83711 93 - 1 AAUUUAUCAAUUAGAGUGUGGCCUUU-GUUAAUUAAAGCCU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGUGGCCAACUAGCAUGGCCAUU----UGGGA .................((((((..(-(((((((.(((((.--------------....))))).)))..((((((......))))))..))))).)))))).----..... ( -29.40) >DroAna_CAF1 80517 110 - 1 AAUUUAUCAAUUAGAGUGUGGCCUUU-GUUAAUUAAAGCCUCGCAGCCACACACAGGCCGCCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAACGUGGCCAUUAUA-UGGGA ......(((......(((((((((((-(.....))))).......)))))))...((((((.(((...(.(((((........))))).)))).))))))......-))).. ( -33.31) >DroPer_CAF1 94573 97 - 1 AAUUUAUCAAUUAGAGUGUGGCCUUUUUUUAAUUAAAGCCU--------------CGCAGUCAUUU-GACUGGCCAAAUUAACGGCCAACUAACGUGGCCAUUAUAUGGGGA ...............(((.(((.(((........)))))).--------------)))((((....-))))..(((...(((.(((((.(....)))))).)))..)))... ( -24.80) >consensus AAUUUAUCAAUUAGAGUGUGGCCUUU_GUUAAUUAAAGCCU______________GGCCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAACGUGGCCAUUA___GGGGA .................((((((...............................................(((((........)))))((....)))))))).......... (-13.28 = -14.00 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:36 2006