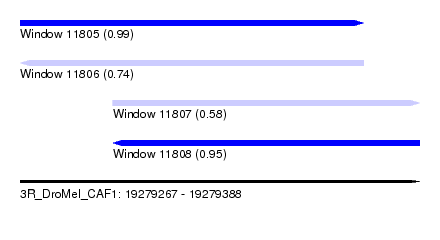
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,279,267 – 19,279,388 |
| Length | 121 |
| Max. P | 0.991357 |

| Location | 19,279,267 – 19,279,371 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -16.38 |
| Energy contribution | -21.62 |
| Covariance contribution | 5.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19279267 104 + 27905053 AUAAGCAAUAGGCCAGGCUAGCUGAAUG--------------AAUGGUUGGAGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCAUGGGCG ..........((((((.((((((.....--------------...))))))....((((.(((....)).).)))-).......))))))..(((....))).(((((.....))))). ( -38.10) >DroGri_CAF1 88558 85 + 1 AUAAGUUGCA----AGCUGAGAAGAA-------------------GGCUGCAGC--GAAGACUGGA--GUUGGCCUGAAACUGGCAGCCCAGUGCGUGAGAAG-UUAAAAGCA------ ....((((((----.(((........-------------------)))))))))--....(((((.--((((.((.......))))))))))).(....)...-.........------ ( -26.50) >DroSec_CAF1 74755 104 + 1 AUAAGCAAUAGACCAGGCUAGCUGAAUG--------------AAUGGAUGGAGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCAUGGGCG ............(((..(((((((....--------------..(......)......))))))).))).(((((-(........)))))).(((....))).(((((.....))))). ( -35.80) >DroSim_CAF1 77015 104 + 1 AUAAGCAAUAGGCCAGGCUAGCUGAAUG--------------AGUGGAUGGAGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCAUGGGCG ..........((((((((((.((..((.--------------.....))...)).))))(((((......)))))-........))))))..(((....))).(((((.....))))). ( -37.20) >DroEre_CAF1 76427 104 + 1 AUAAGCCAUAGGCUUGGCUAGCUGAAUG--------------GGUGGAUGGCGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCACUGCGUGAGCAGGUCCGAAGCAGGGGCG ....(((((..(((...(((.((....)--------------).)))..)))..))))).(((...(((((((((-(........))))).((((....))))))))).)))....... ( -43.10) >DroYak_CAF1 79483 118 + 1 AUAAGCAAUAGGCCAGGCGUGCUGAAUGCAUGCUUGGAGGAUGGGGGAUGUAGGAUGGCGGCUAGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCUGGCCCAAAGCAGGGGCU ............((((((((((.....))))))))))...................((((((((......)))))-.......((((((((((......)))))))...)))....))) ( -43.10) >consensus AUAAGCAAUAGGCCAGGCUAGCUGAAUG______________AGUGGAUGGAGGAUGGCGGCUGGAUGGAUGGCC_AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCAUGGGCG ....((....((((.((((((((.............................(.....)(((((......))))).......))))))))..(((....)))))))....))....... (-16.38 = -21.62 + 5.24)


| Location | 19,279,267 – 19,279,371 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -11.73 |
| Energy contribution | -13.25 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19279267 104 - 27905053 CGCCCAUGCUUUGGGCCUGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCUCCAACCAUU--------------CAUUCAGCUAGCCUGGCCUAUUGCUUAU .......((..((((((.(((.......(((((((......))-))))).......(((..................--------------......))))))..))))))..)).... ( -26.36) >DroGri_CAF1 88558 85 - 1 ------UGCUUUUAA-CUUCUCACGCACUGGGCUGCCAGUUUCAGGCCAAC--UCCAGUCUUC--GCUGCAGCC-------------------UUCUUCUCAGCU----UGCAACUUAU ------.........-........((((((((((((.(((...((((....--....))))..--)))))))))-------------------.......)))..----)))....... ( -18.81) >DroSec_CAF1 74755 104 - 1 CGCCCAUGCUUUGGGCCUGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCUCCAUCCAUU--------------CAUUCAGCUAGCCUGGUCUAUUGCUUAU .......((..((((((.(((.......(((((((......))-))))).......(((..................--------------......))))))..))))))..)).... ( -23.66) >DroSim_CAF1 77015 104 - 1 CGCCCAUGCUUUGGGCCUGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCUCCAUCCACU--------------CAUUCAGCUAGCCUGGCCUAUUGCUUAU .......((..((((((.(((.......(((((((......))-))))).......(((..................--------------......))))))..))))))..)).... ( -26.36) >DroEre_CAF1 76427 104 - 1 CGCCCCUGCUUCGGACCUGCUCACGCAGUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCGCCAUCCACC--------------CAUUCAGCUAGCCAAGCCUAUGGCUUAU .((..(((..(.(((.((((....))))(((((((......))-)))))))))..)))..))...............--------------.........(((((......)))))... ( -26.10) >DroYak_CAF1 79483 118 - 1 AGCCCCUGCUUUGGGCCAGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCUAGCCGCCAUCCUACAUCCCCCAUCCUCCAAGCAUGCAUUCAGCACGCCUGGCCUAUUGCUUAU .......((..((((((((.........(((((((......))-))))).....................................((.(((.....))).))))))))))..)).... ( -32.40) >consensus CGCCCAUGCUUUGGGCCUGCUCACGCACUGGCCAGCUUCUUUU_GGCCAUCCAUCCAGCCGCCAUCCUCCAUCCACU______________CAUUCAGCUAGCCUGGCCUAUUGCUUAU .......((..((((((.((....))..(((((...........)))))........................................................))))))..)).... (-11.73 = -13.25 + 1.52)


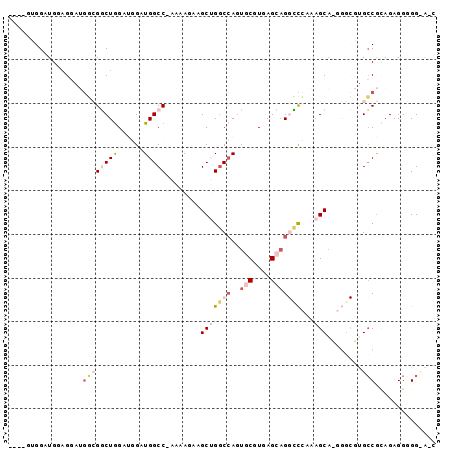
| Location | 19,279,295 – 19,279,388 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.13 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -9.15 |
| Energy contribution | -11.27 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
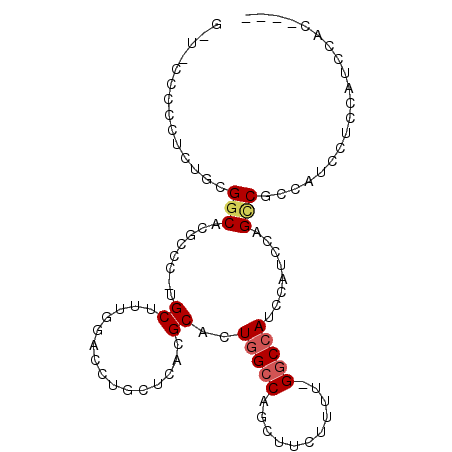
>3R_DroMel_CAF1 19279295 93 + 27905053 ---AAUGGUUGGAGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCAUGGGCGUGCCGAAGAGG-GGUAUC ---..((((..(........(((((......)))))-........)..)))).(((....))).(((((.....)))))(((((.......-))))). ( -31.99) >DroVir_CAF1 94518 80 + 1 ------GGCUGCAGC--UUAGACUGGG--GUUGGCCCGAAACUGGCAGGCGAGUGCGUGAGAAG-UUAAAAGCC-------GACACAGAGGUGGCGGC ------.(((((.((--((...(((..--((((((....((((..((.((....)).))...))-))....)))-------))).))))))).))))) ( -29.50) >DroGri_CAF1 88580 80 + 1 ------GGCUGCAGC--GAAGACUGGA--GUUGGCCUGAAACUGGCAGCCCAGUGCGUGAGAAG-UUAAAAGCA-------GCCGCAGAGGUGGCGCU ------((((...((--(...(((((.--((((.((.......))))))))))).)))....))-))...(((.-------(((((....)))))))) ( -29.70) >DroSim_CAF1 77043 94 + 1 ---AGUGGAUGGAGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCAUGGGCGUGCCGUAGAGGGGGUAUC ---....((((.......(((((........(((((-(........)))))).(((....))).(((((.....)))))..)))))........)))) ( -32.36) >DroEre_CAF1 76455 91 + 1 ---GGUGGAUGGCGGAUGGCGGCUGGAUGGAUGGCC-AAAAGAAGCUGGCCACUGCGUGAGCAGGUCCGAAGCAGGGGCGUGCCGCAGAGGGGG-A-- ---........(((((((.(.(((...(((((((((-(........))))).((((....))))))))).)))...).))).))))........-.-- ( -37.40) >DroYak_CAF1 79522 94 + 1 GAUGGGGGAUGUAGGAUGGCGGCUAGAUGGAUGGCC-AAAAGAAGCUGGCCAGUGCGUGAGCUGGCCCAAAGCAGGGGCUUGCCGUAGAGGGGG-A-- ...............((((((((((......)))))-.....((((((((((((......))))))).........))))))))))........-.-- ( -30.60) >consensus ____GUGGAUGGAGGAUGGCGGCUGGAUGGAUGGCC_AAAAGAAGCUGGCCAGUGCGUGAGCAGGCCCAAAGCA_GGGCGUGCCGCAGAGGGGG_A_C .................(((.(((.(.....(((((...........))))).....).)))..((((.......))))..))).............. ( -9.15 = -11.27 + 2.11)

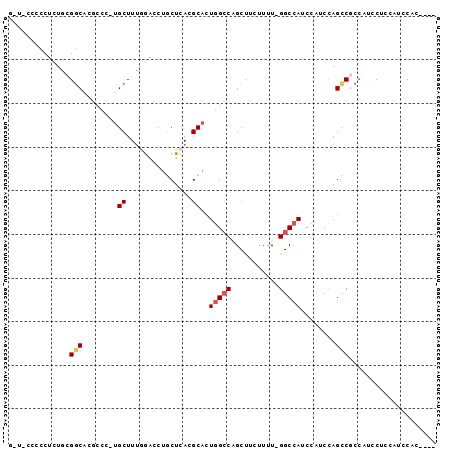
| Location | 19,279,295 – 19,279,388 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.13 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -7.77 |
| Energy contribution | -8.16 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19279295 93 - 27905053 GAUACC-CCUCUUCGGCACGCCCAUGCUUUGGGCCUGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCUCCAACCAUU--- ......-.......(((..(((((.....)))))..(((.......(((((((......))-))))).......))).)))..............--- ( -26.44) >DroVir_CAF1 94518 80 - 1 GCCGCCACCUCUGUGUC-------GGCUUUUAA-CUUCUCACGCACUCGCCUGCCAGUUUCGGGCCAAC--CCCAGUCUAA--GCUGCAGCC------ ((((.(((....))).)-------)))......-........((....))((((.(((((.((((....--....))))))--)))))))..------ ( -21.20) >DroGri_CAF1 88580 80 - 1 AGCGCCACCUCUGCGGC-------UGCUUUUAA-CUUCUCACGCACUGGGCUGCCAGUUUCAGGCCAAC--UCCAGUCUUC--GCUGCAGCC------ ((((((........)))-------.))).....-..............((((((.(((...((((....--....))))..--)))))))))------ ( -21.30) >DroSim_CAF1 77043 94 - 1 GAUACCCCCUCUACGGCACGCCCAUGCUUUGGGCCUGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCUCCAUCCACU--- .............((((..(((((.....))))).(((....))).(((((((......))-)))))........))))................--- ( -26.80) >DroEre_CAF1 76455 91 - 1 --U-CCCCCUCUGCGGCACGCCCCUGCUUCGGACCUGCUCACGCAGUGGCCAGCUUCUUUU-GGCCAUCCAUCCAGCCGCCAUCCGCCAUCCACC--- --.-........(((((..((....))...(((.((((....))))(((((((......))-)))))))).....)))))...............--- ( -28.30) >DroYak_CAF1 79522 94 - 1 --U-CCCCCUCUACGGCAAGCCCCUGCUUUGGGCCAGCUCACGCACUGGCCAGCUUCUUUU-GGCCAUCCAUCUAGCCGCCAUCCUACAUCCCCCAUC --.-..........(((..((((.......))))..(((.......(((((((......))-))))).......))).)))................. ( -23.84) >consensus G_U_CCCCCUCUGCGGCACGCCC_UGCUUUGGACCUGCUCACGCACUGGCCAGCUUCUUUU_GGCCAUCCAUCCAGCCGCCAUCCUCCAUCCAC____ ..............(((........((...............))..(((((...........)))))........))).................... ( -7.77 = -8.16 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:30 2006