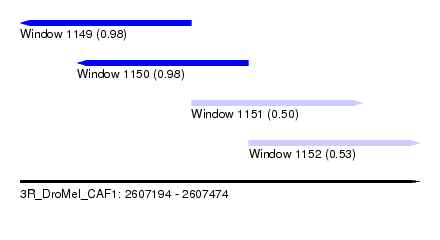
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,607,194 – 2,607,474 |
| Length | 280 |
| Max. P | 0.976226 |

| Location | 2,607,194 – 2,607,314 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.23 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2607194 120 - 27905053 UUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUAGGGGAUUGUAUUUGUGGAAAGUUCGGGGAUGGUUUCCGACGUGACAUUUGUCCUGUGUGACGGGGAGUUUCACUAAAGUAAA ...(((((((((.....)))))))))((((((((((((((........))))))))))))))..((.(((.((((..(((.((((.......)))).))).))))..)))))........ ( -38.30) >DroSim_CAF1 71200 120 - 1 UUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUGGGGGAUUGUAUUUGUGGAAAGUUCGGGGAUGGUUUUCGACGUGACAUGUGCCCUGUGUGACGGGGAGUUUCACUAAAGUAAA ...(((((((((.....)))))))))(((((((((((..(........)..)))))))))))(..((......))..)((((.((...(((((.....))))).)).))))......... ( -38.80) >DroEre_CAF1 76080 120 - 1 UUAUUAGCAGCUUAGGCAGCUGUUAAGAGCUUUUCAUAGGGGAUUGUAUUUGUGGAAAGUUUGGGGAUGGUUUCCGACGUGACAUUUGUCCUCUGUGACGGGGAGUUUCAAUAAAGUAAA ...(((((((((.....)))))))))(((((((((((((((((((((...((((((((.(........).))))).)))..)))...)))))))))))...)))))))............ ( -37.00) >DroYak_CAF1 74666 120 - 1 UUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUAGGGGAUUCUAUUUGUAGAAAGUUCGGGGAUGGUUUCCGACGUGACAUUUGCCCUCUGGAAUGGGGAGUUUCACUAAAGUAAA ...(((((((((.....)))))))))((((((..((((((((.(((((....))))).(((((.(((.....)))..)).)))......)))))...)))..))))))............ ( -34.40) >consensus UUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUAGGGGAUUGUAUUUGUGGAAAGUUCGGGGAUGGUUUCCGACGUGACAUUUGCCCUCUGUGACGGGGAGUUUCACUAAAGUAAA ...(((((((((.....)))))))))((((((((((((((........)))))))))))))).((((....))))...((((.((...(((((.....))))).)).))))......... (-34.22 = -34.23 + 0.00)

| Location | 2,607,234 – 2,607,354 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -41.40 |
| Energy contribution | -41.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2607234 120 - 27905053 CCACAUCCGCACUGACCAAUUCCUGGCCUGUGUCCAUUCGUUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUAGGGGAUUGUAUUUGUGGAAAGUUCGGGGAUGGUUUCCGACGU ...(((((((((.(.(((.....))).).))))((........(((((((((.....)))))))))((((((((((((((........)))))))))))))))))))))........... ( -41.30) >DroSim_CAF1 71240 120 - 1 CCACAUCCGCACUGACCAAUUCCUGGCCUGUGUCCAUUCGUUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUGGGGGAUUGUAUUUGUGGAAAGUUCGGGGAUGGUUUUCGACGU ...(((((((((.(.(((.....))).).))))((........(((((((((.....)))))))))(((((((((((..(........)..))))))))))))))))))........... ( -40.80) >DroEre_CAF1 76120 120 - 1 CCACAUCCACACUGACCAAUUCCUGGCCGGUGUCCAUUCGUUAUUAGCAGCUUAGGCAGCUGUUAAGAGCUUUUCAUAGGGGAUUGUAUUUGUGGAAAGUUUGGGGAUGGUUUCCGACGU ...(((((((((((.(((.....))).))))))((........(((((((((.....)))))))))((((((((((((((........)))))))))))))))))))))........... ( -42.00) >DroYak_CAF1 74706 120 - 1 CCACAUCCGCACUGACCAAUUCCUGGUCGGUGUCCAUUCGUUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUAGGGGAUUCUAUUUGUAGAAAGUUCGGGGAUGGUUUCCGACGU ...(((((((((((((((.....))))))))))((........(((((((((.....)))))))))(((((((..(((((........)))))..))))))))))))))........... ( -45.30) >consensus CCACAUCCGCACUGACCAAUUCCUGGCCGGUGUCCAUUCGUUAUUAGCAGCUUAGGCAGCUGUUGAGAGCUUUUCAUAGGGGAUUGUAUUUGUGGAAAGUUCGGGGAUGGUUUCCGACGU ...(((((((((((.(((.....))).))))))((........(((((((((.....)))))))))((((((((((((((........)))))))))))))))))))))........... (-41.40 = -41.40 + -0.00)

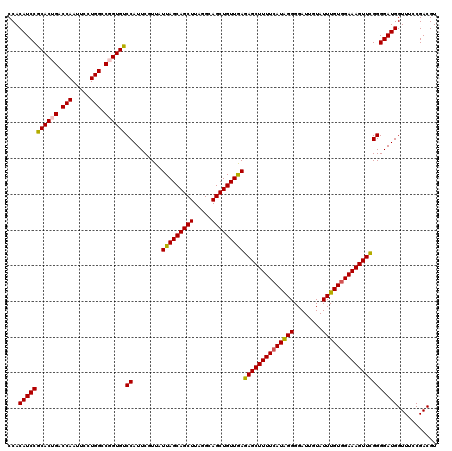
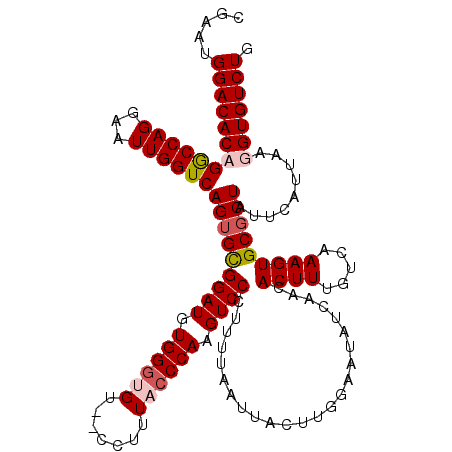
| Location | 2,607,314 – 2,607,434 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -31.12 |
| Energy contribution | -32.25 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2607314 120 + 27905053 CGAAUGGACACAGGCCAGGAAUUGGUCAGUGCGGAUGUGGGUGUCUCCUUUACCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUG .....((((((.((((((...))))))(((((((((.((((((.......)))))).))))........................((((.....)))))))))..........)))))). ( -35.20) >DroSim_CAF1 71320 118 + 1 CGAAUGGACACAGGCCAGGAAUUGGUCAGUGCGGAUGUGGGUGU--CCUUUACCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUG .....((((((.((((((...))))))(((((((((.((((((.--....)))))).))))........................((((.....)))))))))..........)))))). ( -35.50) >DroEre_CAF1 76200 118 + 1 CGAAUGGACACCGGCCAGGAAUUGGUCAGUGUGGAUGUGGGUGU--CCUUUUCCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUG .....(((((((((((((...))))))((((.((((.((((...--......)))).)))).......................(((((.....))))))))).........))))))). ( -33.40) >DroYak_CAF1 74786 117 + 1 CGAAUGGACACCGACCAGGAAUUGGUCAGUGCGGAUGUGGAUGU--CCUUUUCCCAAGUCCA-UUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCUCUAUUCAUUAAGGUGUCUG .....(((((((((((((...))))))((((..(((((((....--)).....((((((...-.......)))))).)))))..))))........................))))))). ( -33.70) >consensus CGAAUGGACACAGGCCAGGAAUUGGUCAGUGCGGAUGUGGGUGU__CCUUUACCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUG .....(((((((((((((...))))))(((((((((.((((((.......)))))).))))........................((((.....))))))))).........))))))). (-31.12 = -32.25 + 1.12)

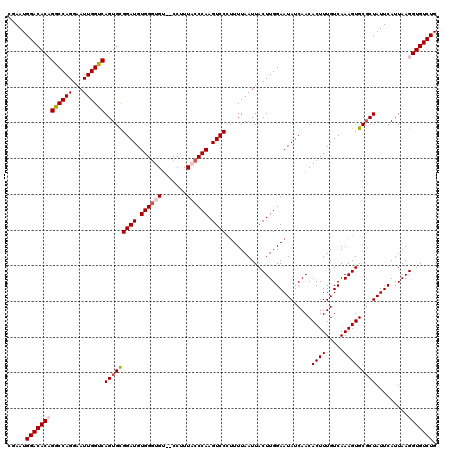
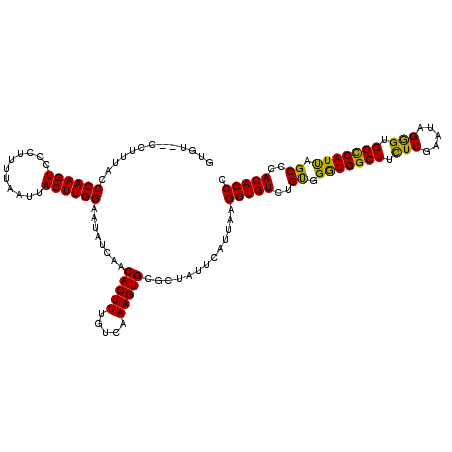
| Location | 2,607,354 – 2,607,474 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -30.88 |
| Energy contribution | -29.75 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.99 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2607354 120 + 27905053 GUGUCUCCUUUACCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUGUGGGUGGCUUUUCGAAUAGGGUGGCCAUUAGCCCACACCG (((..........((((((...........))))))........(((((.....))))))))..........(((((..((..((((((.(((.....))).))))))..))..))))). ( -30.20) >DroSim_CAF1 71360 118 + 1 GUGU--CCUUUACCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUGUGGGUGGCUUCUCGAAUGGGGUGGCCAUUAGCCCACACCG (((.--.((((..((((((...........)))))).....(((....)))..))))..)))..........(((((..((..((((((.(((.....))).))))))..))..))))). ( -32.70) >DroEre_CAF1 76240 118 + 1 GUGU--CCUUUUCCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUGCGGAUGGCUUCUCGAAUGGGGUGGUCAUUAGCCCACACCC (((.--.((((..((((((...........)))))).....(((....)))..))))..)))..........(((((..((.(((((((.(((.....))).))))))).))..))))). ( -30.00) >DroYak_CAF1 74826 117 + 1 AUGU--CCUUUUCCCAAGUCCA-UUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCUCUAUUCAUUAAGGUGUCUGUGGGUGGCUUCUCGAAUAGAGUGGCCAUCAGCCCACACCC ....--.......((((((...-.......))))))........(((((.....))))).............(((((..((.(((((((.(((.....))).))))))).))..))))). ( -32.30) >consensus GUGU__CCUUUACCCAAGUCCCUUUUAAUUACUUGGAAUAUCAACACUUUGUCAAAGUGCGCUAUUCAUUAAGGUGUCUGUGGGUGGCUUCUCGAAUAGGGUGGCCAUUAGCCCACACCC .............((((((...........))))))........(((((.....))))).............(((((..((.(((((((.(((.....))).))))))).))..))))). (-30.88 = -29.75 + -1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:27 2006