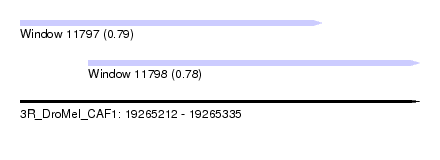
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,265,212 – 19,265,335 |
| Length | 123 |
| Max. P | 0.793235 |

| Location | 19,265,212 – 19,265,305 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -14.43 |
| Energy contribution | -14.35 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
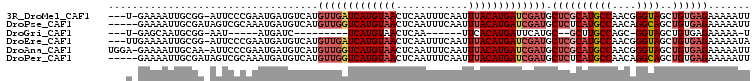
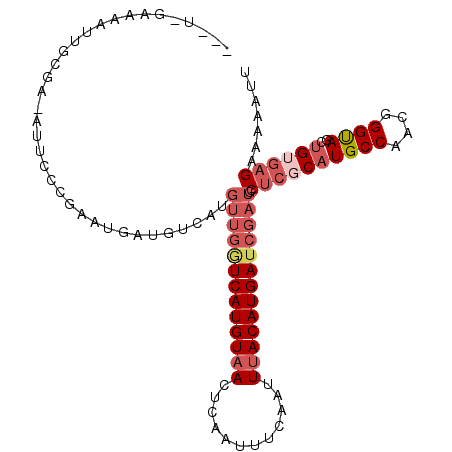
>3R_DroMel_CAF1 19265212 93 + 27905053 GUGCAAUUUCUCUAUUAUCGA-------U-GAAAAUUGCGG-AUUCCCGAAUGAUGUCAUGUUGAUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC ..((................(-------(-((..(((.(((-....))))))....))))(((((((((((((............))))))))))))))).. ( -21.50) >DroPse_CAF1 75854 79 + 1 --------------AGAUCGA---------GAAAAUUGCGAUAGUCGCAAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC --------------.....((---------(....(((((.....)))))..........(((((((((((((............))))))))))))).))) ( -20.40) >DroSim_CAF1 61215 93 + 1 GUGCAGUUUUUCUAUUAUCGA-------U-GAAAAUUACGG-AUUCCCGAAUGAUGUCAUGUUGAUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC ..((................(-------(-((..(((.(((-....))))))....))))(((((((((((((............))))))))))))))).. ( -21.50) >DroEre_CAF1 62070 94 + 1 GUGCAGUUUCUCUAUUAUCGA-------UUGAAAAUUGCGG-AUUCCCGAAUGAUGUCAUGUUGAUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC ..(((((((.((.........-------..)))))))))((-....))..(((....)))(((((((((((((............))))))))))))).... ( -20.90) >DroAna_CAF1 62797 98 + 1 --GGAAGUGCUCCAUUACCAAAGUUUGGA-GAAAAUUGCAA-AUUCCCGAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC --(((.....))).....((..((((((.-(((........-.)))))))))..))....(((((((((((((............))))))))))))).... ( -21.60) >DroPer_CAF1 75961 79 + 1 --------------AGAUCGA---------GAAAAUUGCGAUAGUCGCAAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC --------------.....((---------(....(((((.....)))))..........(((((((((((((............))))))))))))).))) ( -20.40) >consensus __GCA_UUUCUCUAUUAUCGA_______U_GAAAAUUGCGA_AUUCCCGAAUGAUGUCAUGUUGAUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUC .....................................((...........(((....)))(((((((((((((............))))))))))))))).. (-14.43 = -14.35 + -0.08)

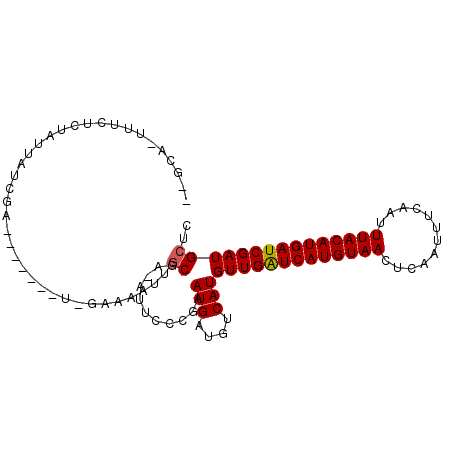
| Location | 19,265,233 – 19,265,335 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -18.66 |
| Energy contribution | -20.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19265233 102 + 27905053 ---U-GAAAAUUGCGG-AUUCCCGAAUGAUGUCAUGUUGAUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUCGCAUGCCAACGGGUAGCUGUGAGAAAAAUU ---(-((..(((.(((-....))))))....))).(((((((((((((............))))))))))))).((((((((((....))))..))))))....... ( -28.80) >DroPse_CAF1 75861 102 + 1 -----GAAAAUUGCGAUAGUCGCAAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUCUCAUGCCAACAGGCAGCUGUGAGAAAAAUU -----.....(((((.....)))))(((....)))(((((((((((((............)))))))))))))..(((((((((....))))....)))))...... ( -28.60) >DroGri_CAF1 74617 78 + 1 ---U-GAGCAAUGCGG-AAU-----AUGAUC---------UCAUGUAACUCAA------UUCACAUGAUUCAUGC--GCUUGCCAGC-GGUAGCUGUGAGAAAAA-U ---.-..((((.(((.-..(-----((((..---------((((((.......------...)))))).))))))--))))))((((-....)))).........-. ( -20.00) >DroEre_CAF1 62091 103 + 1 ---UUGAAAAUUGCGG-AUUCCCGAAUGAUGUCAUGUUGAUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUCGCAUGCCAACGGGUAGCUGUGAGAAAAAUA ---.(((..(((.(((-....))))))....))).(((((((((((((............))))))))))))).((((((((((....))))..))))))....... ( -29.00) >DroAna_CAF1 62820 105 + 1 UGGA-GAAAAUUGCAA-AUUCCCGAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUCGCAUGCCAACGGGUAGCUGUGAGAAAAAUU .((.-(((........-.)))))..(((....)))(((((((((((((............))))))))))))).((((((((((....))))..))))))....... ( -25.50) >DroPer_CAF1 75968 102 + 1 -----GAAAAUUGCGAUAGUCGCAAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUCUCAUGCCAACAGGCAGCUGUGAGAAAAAUU -----.....(((((.....)))))(((....)))(((((((((((((............)))))))))))))..(((((((((....))))....)))))...... ( -28.60) >consensus ___U_GAAAAUUGCGA_AUUCCCGAAUGAUGUCAUGUUGGUCAUGUAACUCAAUUUCAAUUUACAUGAUCGAUGCUCGCAUGCCAACGGGUAGCUGUGAGAAAAAUU ...................................(((((((((((((............))))))))))))).((((((((((....))))..))))))....... (-18.66 = -20.10 + 1.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:21 2006