| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,233,241 – 19,233,343 |
| Length | 102 |
| Max. P | 0.758639 |

| Location | 19,233,241 – 19,233,343 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
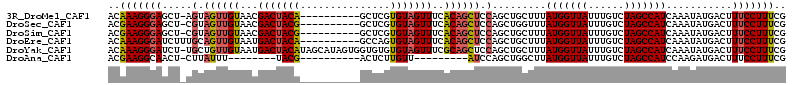
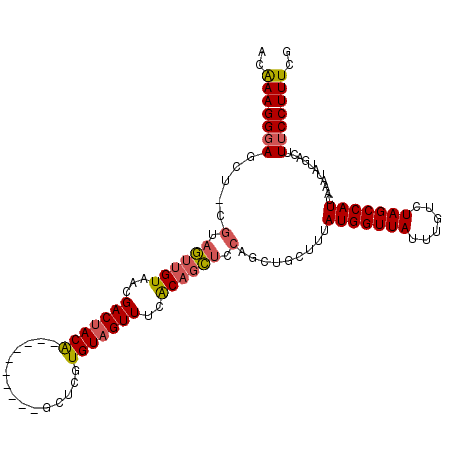
>3R_DroMel_CAF1 19233241 102 + 27905053 ACAAAGGGAGCU-AGUAGUUGUAACGACUACA----------GCUCGUGUAGUUUCACAGCUCCAGCUGCUUUAUGGUUAUUUGUCUAGCCAUCAAAUAUGACUUUCCUUUCG ..((((((((((-.(((((((...))))))))----------))((((((.......((((....))))....(((((((......)))))))...))))))..))))))).. ( -29.60) >DroSec_CAF1 29029 102 + 1 ACGAAGGGAGCU-CGUAGUUGUAACGACUACG----------GCUCGUGUAGUUUCACAGCUCCAGCUGGUUUAUGGUUAUUUGUCUAGCCAUCAAAUAUGACUUUCCUUUCG .((((((((((.-((((((((...))))))))----------))((((((.......((((....))))....(((((((......)))))))...))))))...)))))))) ( -36.70) >DroSim_CAF1 29296 102 + 1 ACGAAGGGAGCU-CGUAGUUGUAACGACUACG----------GCUCGUGUAGUUUCACAGCUCCAGCUGCUUUAUGGUUAUUUGUCUAGCCAUCAAAUAUGACUUUCCUUUCG .((((((((((.-((((((((...))))))))----------))((((((.......((((....))))....(((((((......)))))))...))))))...)))))))) ( -35.80) >DroEre_CAF1 29952 103 + 1 ACAAAGGGAUCUUUGCAGUUGUAAUGACUACA----------GCCAGUGUAGUUUCACAGCUCCAGCUGCUUUAUGGUUAUUUGUCUAGCCAUCAAAUAUGACUUUCCUUUCG ..(((((((.....(((((((....(((((((----------.....))))))).........)))))))...(((((((......)))))))...........))))))).. ( -25.72) >DroYak_CAF1 30497 112 + 1 ACAAAGGGAUCU-UGCUGUUGUAAUGACUACAUAGCAUAGUGGUGUGUGUAGUUUCGCAGCUCCAGCUGCUUUAUGGUUAUUUGUCUAGCCAUCAAAUAUGACUUUCCUUUCG ..(((((((...-((((((.(((.....))))))))).(((.((((.((.......(((((....)))))...(((((((......))))))))).)))).)))))))))).. ( -31.60) >DroAna_CAF1 30029 85 + 1 ACGAAGGCAACU-CUUAUUU--------UACG----------ACUCUUGUU---------AUCCAGCUGGCUUAUGGUUAUUUGUCUAGCCAUCCAAGAUGACUUUCCUUUCG .(((((....).-.......--------....----------......(((---------(((....(((...(((((((......)))))))))).))))))......)))) ( -15.50) >consensus ACAAAGGGAGCU_CGUAGUUGUAACGACUACA__________GCUCGUGUAGUUUCACAGCUCCAGCUGCUUUAUGGUUAUUUGUCUAGCCAUCAAAUAUGACUUUCCUUUCG ..(((((((.....(.((((((...(((((((...............)))))))..)))))).).........(((((((......)))))))...........))))))).. (-18.14 = -18.46 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:07 2006