| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,224,196 – 19,224,290 |
| Length | 94 |
| Max. P | 0.854064 |

| Location | 19,224,196 – 19,224,290 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19224196 94 + 27905053 --------UUCGGCCAAAAGGUGCCAAUGUGGCAAAUGUUUUGUGCUUAACAGUUUUAUGGCUGCCAUAAGUUGGC-CGGAGUGUGGUAAAUGCUGCUGCCAC --------(((((((((..(....)..(((((((.(((..((((.....))))...)))...)))))))..)))))-))))..((((((........)))))) ( -31.00) >DroVir_CAF1 39206 85 + 1 ---------UUGGCCAAAAGGUGCCAAUGUGGCAAAUGUUGUGCGCUUAACAUUUUUAUGACCACCAUAAGUCGUCUUAGAAUGUGGUAAAUGU--------- ---------...(((...((((((((((((.....)))))).)))))).(((((((...(((.((.....)).)))..))))))))))......--------- ( -23.00) >DroPse_CAF1 33167 99 + 1 GUUUUUUCGUUGGCCAAAAGGUGCCAAUGUGGCAAAUGUUUUGUGCUUAACAUUUUUAUGGCUGCCAUAAGUUGGC-UAGAAUGUGGUAAAUGCUGCAGG--- .....(((...(((((......(((.....)))(((((((........)))))))...)))))((((.....))))-..)))((..((....))..))..--- ( -26.00) >DroGri_CAF1 35704 94 + 1 ---------UUGGCAUAAAGGUGCCAAUGUGGCAAAUGUUGUGAGCUUAACAUUUUUAUGGCUAACAUAAGUCGACUUAGAAUGUAGUAAAUGUGGCAGACUC ---------(((((((....)))))))(((.((((((((((......)))))))......((((...((((....)))).....))))...))).)))..... ( -22.90) >DroAna_CAF1 20504 94 + 1 --------UUCGGCCAAAAGGUGCCAAUGUGGCAAAUGUUUUGUGCUUAACAUUUUUAUGGCUGCCAUAAGUUGGC-CAGAGUGUGGUAAAUACUGCAGUUAC --------...((((((....((((.....))))((((((........))))))(((((((...))))))))))))-).((.((..((....))..)).)).. ( -28.00) >DroPer_CAF1 33216 99 + 1 GUUUUUUUGUUGGCCAAAAGGUGCCAAUGUGGCAAAUGUUUUGUGCUUAACAUUUUUAUGGCUGCCAUAAGUUGGC-UAGAAUGUGGUAAAUGCUGCAGG--- .........((((((((....((((.....))))((((((........))))))(((((((...))))))))))))-)))..((..((....))..))..--- ( -25.60) >consensus _________UUGGCCAAAAGGUGCCAAUGUGGCAAAUGUUUUGUGCUUAACAUUUUUAUGGCUGCCAUAAGUUGGC_UAGAAUGUGGUAAAUGCUGCAGG___ ...........(((((......(((.....)))(((((((........)))))))...)))))((((((..((......)).))))))............... (-16.36 = -16.78 + 0.42)

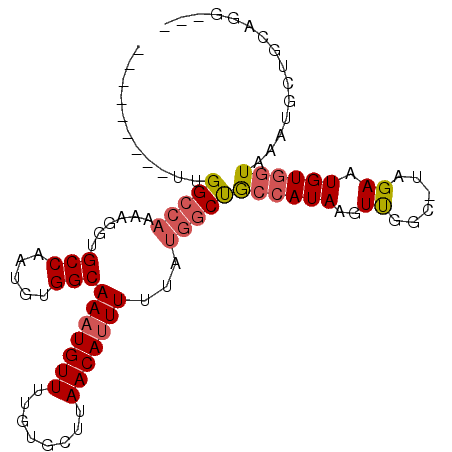
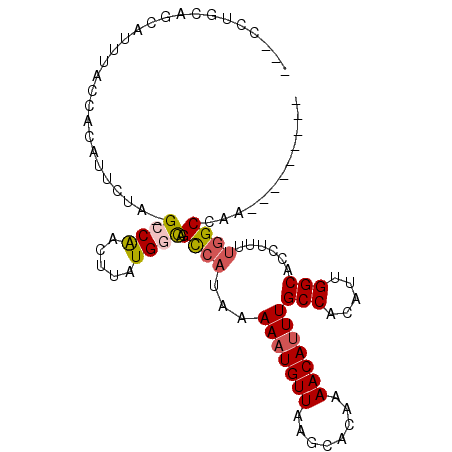
| Location | 19,224,196 – 19,224,290 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -19.72 |
| Consensus MFE | -13.60 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19224196 94 - 27905053 GUGGCAGCAGCAUUUACCACACUCCG-GCCAACUUAUGGCAGCCAUAAAACUGUUAAGCACAAAACAUUUGCCACAUUGGCACCUUUUGGCCGAA-------- ((((............))))....((-(((((....((((((........)))))).............((((.....))))....)))))))..-------- ( -25.60) >DroVir_CAF1 39206 85 - 1 ---------ACAUUUACCACAUUCUAAGACGACUUAUGGUGGUCAUAAAAAUGUUAAGCGCACAACAUUUGCCACAUUGGCACCUUUUGGCCAA--------- ---------......((((.....((((....))))))))(((((...(((((((........)))))))(((.....)))......)))))..--------- ( -17.50) >DroPse_CAF1 33167 99 - 1 ---CCUGCAGCAUUUACCACAUUCUA-GCCAACUUAUGGCAGCCAUAAAAAUGUUAAGCACAAAACAUUUGCCACAUUGGCACCUUUUGGCCAACGAAAAAAC ---..................(((..-((((.....)))).((((...(((((((........)))))))(((.....)))......))))....)))..... ( -19.80) >DroGri_CAF1 35704 94 - 1 GAGUCUGCCACAUUUACUACAUUCUAAGUCGACUUAUGUUAGCCAUAAAAAUGUUAAGCUCACAACAUUUGCCACAUUGGCACCUUUAUGCCAA--------- (((.((...(((((((((........)))....(((((.....)))))))))))..)))))...............((((((......))))))--------- ( -14.90) >DroAna_CAF1 20504 94 - 1 GUAACUGCAGUAUUUACCACACUCUG-GCCAACUUAUGGCAGCCAUAAAAAUGUUAAGCACAAAACAUUUGCCACAUUGGCACCUUUUGGCCGAA-------- ........................((-(((((.((((((...))))))(((((((........)))))))(((.....))).....)))))))..-------- ( -21.50) >DroPer_CAF1 33216 99 - 1 ---CCUGCAGCAUUUACCACAUUCUA-GCCAACUUAUGGCAGCCAUAAAAAUGUUAAGCACAAAACAUUUGCCACAUUGGCACCUUUUGGCCAACAAAAAAAC ---.......................-((((.....)))).((((...(((((((........)))))))(((.....)))......))))............ ( -19.00) >consensus ___CCUGCAGCAUUUACCACAUUCUA_GCCAACUUAUGGCAGCCAUAAAAAUGUUAAGCACAAAACAUUUGCCACAUUGGCACCUUUUGGCCAA_________ ...........................((((.....)))).((((...(((((((........)))))))(((.....)))......))))............ (-13.60 = -13.85 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:59 2006