
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,193,803 – 19,193,918 |
| Length | 115 |
| Max. P | 0.985881 |

| Location | 19,193,803 – 19,193,918 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -41.75 |
| Consensus MFE | -35.00 |
| Energy contribution | -36.50 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
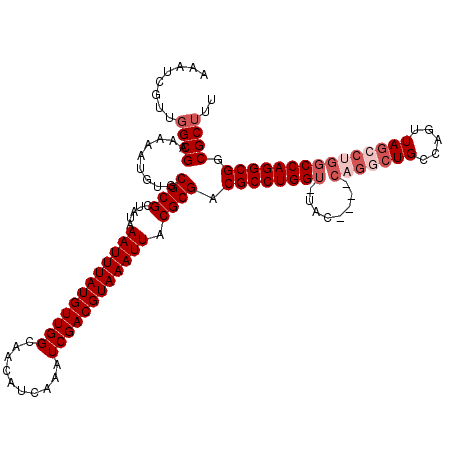
>3R_DroMel_CAF1 19193803 115 + 27905053 AAAUCGUUGGCGAAAAAUGUGCGCGCUAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGU-UAC----CAGGCUGCCAGUUAGCCUGGCCAGGCGGCGCUUU ...(((....))).......((((((....((((((((((((..........))))))))))))..))..(((((((.-..(----(((((((.....)))))))))))))))))))... ( -46.20) >DroSec_CAF1 5776 119 + 1 AAAUCGUUGGCGAAAAAUGUUCGCGCUAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGU-UACCAACCAGACUGCCAGUUAGCCUGGCCAGGCGGCGCUUU .........(((((.....)))))......((((((((((((..........))))))))))))..(((.((((((((-(........))).(((((.....))))))))))).)))... ( -37.80) >DroSim_CAF1 6174 119 + 1 AAAUCGUUGGCGAAAAAUGUGCGCGCUAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGU-UACCAACCAGGCUGCCAGUUAGCCUGGCCAGGCGGCGCUUU ...(((....))).......((((((....((((((((((((..........))))))))))))..))..(((((((.-......((((((((.....)))))))))))))))))))... ( -44.81) >DroEre_CAF1 7122 115 + 1 AAAUCGUUGGCGAAAAAUGUGCGCGCUAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGU-UAC----CAGGCUGCCAGUUAGCCCGGCCAGGCGGCGCUUU ...(((....))).......((((((....((((((((((((..........))))))))))))..))..(((((((.-..(----(.(((((.....))))).)))))))))))))... ( -41.60) >DroYak_CAF1 9737 116 + 1 AAAUCGUUGGCGAAAAAUGUGCGCGCUAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGUUUAC----CAGGCUGCCAGUUAGCCUGGCCAGGCGGCGCUUU ...(((....))).......((((((....((((((((((((..........))))))))))))..))..(((((((....(----(((((((.....)))))))))))))))))))... ( -45.80) >DroAna_CAF1 11907 101 + 1 ---------GCGAAAAAUGUGCGCGCGAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGU-UAC----CAGGCUGCCAGUUAU-----CCAGGCGGCGCUUU ---------...........(((((((...((((((((((((..........)))))))))))).)))..(((((((.-.((----..((...)).))...-----)))))))))))... ( -34.30) >consensus AAAUCGUUGGCGAAAAAUGUGCGCGCUAUAAAUUUAUGUUGGCAACAUCAAAUCGACGUAAAUUACGCGACGCCUGGU_UAC____CAGGCUGCCAGUUAGCCUGGCCAGGCGGCGCUUU ........((((.........((((.....((((((((((((..........)))))))))))).)))).((((((((........(((((((.....))))))))))))))).)))).. (-35.00 = -36.50 + 1.50)

| Location | 19,193,803 – 19,193,918 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -37.91 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.83 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19193803 115 - 27905053 AAAGCGCCGCCUGGCCAGGCUAACUGGCAGCCUG----GUA-ACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUAGCGCGCACAUUUUUCGCCAACGAUUU ...(((.((((((((((((((.......))))))----)..-.))))))).)))........((((((...(((((((((..............).))).)))))..)))...))).... ( -39.54) >DroSec_CAF1 5776 119 - 1 AAAGCGCCGCCUGGCCAGGCUAACUGGCAGUCUGGUUGGUA-ACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUAGCGCGAACAUUUUUCGCCAACGAUUU ...(((.(((((((....((((((..(....)..)))))).-.))))))).)))((.....(((((.....))))).....))...........(.(((((.....))))))........ ( -38.10) >DroSim_CAF1 6174 119 - 1 AAAGCGCCGCCUGGCCAGGCUAACUGGCAGCCUGGUUGGUA-ACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUAGCGCGCACAUUUUUCGCCAACGAUUU ...(((((((((....))))....((((((((((((.....-)))))))((((((((...)))).)))).......))))).............).)))).......(((....)))... ( -40.00) >DroEre_CAF1 7122 115 - 1 AAAGCGCCGCCUGGCCGGGCUAACUGGCAGCCUG----GUA-ACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUAGCGCGCACAUUUUUCGCCAACGAUUU ...(((.((((((((((((((.......))))))----)..-.))))))).)))........((((((...(((((((((..............).))).)))))..)))...))).... ( -38.84) >DroYak_CAF1 9737 116 - 1 AAAGCGCCGCCUGGCCAGGCUAACUGGCAGCCUG----GUAAACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUAGCGCGCACAUUUUUCGCCAACGAUUU ...(((((((((....))))....((((((((((----(....))))))((((((((...)))).)))).......))))).............).)))).......(((....)))... ( -39.40) >DroAna_CAF1 11907 101 - 1 AAAGCGCCGCCUGG-----AUAACUGGCAGCCUG----GUA-ACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUCGCGCGCACAUUUUUCGC--------- ...((((.((.(((-----((...((((((((((----(..-.))))))((((((((...)))).)))).......)))))......)))))..))))))...........--------- ( -31.60) >consensus AAAGCGCCGCCUGGCCAGGCUAACUGGCAGCCUG____GUA_ACCAGGCGUCGCGUAAUUUACGUCGAUUUGAUGUUGCCAACAUAAAUUUAUAGCGCGCACAUUUUUCGCCAACGAUUU ...((((.((((....))))....((((((((((..........)))))((((((((...)))).)))).......))))).............))))...................... (-30.42 = -30.83 + 0.42)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:42 2006