
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
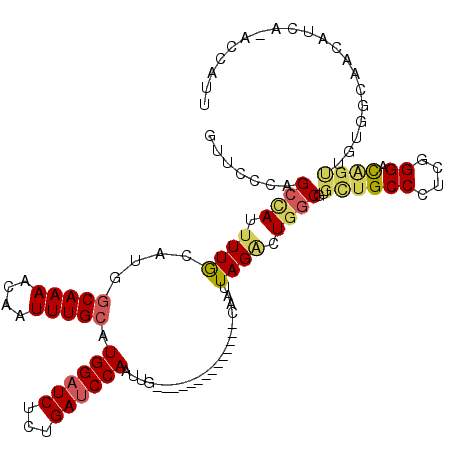
| Location | 19,192,895 – 19,193,000 |
| Length | 105 |
| Max. P | 0.838059 |

| Location | 19,192,895 – 19,193,000 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19192895 105 + 27905053 GUUCCCAGACAUUUUGCAUGGCAAAACAAUUUGCAUGGAUCUCUGAUCCAAUUG--------------CAAUUAGACUGGCGUGCUGCCCUCGGGACAGUUGUGGCAACAUCA-ACCAUU (((((.((.......(((((.((.......(((((((((((...))))))..))--------------)))......)).)))))....)).))))).((((((....)).))-)).... ( -32.42) >DroSec_CAF1 4885 106 + 1 GUUCCCAGCCAUUUUGCAGCGCAAAACAAUUUGCAUGGAUCUCUGAUCCAAUUG--------------CAAUUAGACUGGCGUGCUGCCCUCGGGACAGUUGUGGCAACAUCAAACCAUU (((.(((((((..((((((.(((((....))))).((((((...)))))).)))--------------)))......))))..((((((....)).))))..))).)))........... ( -32.40) >DroSim_CAF1 5261 106 + 1 GUUCCCAGCCAUUUUGCAGCGCAAAACAAUUUGCAUGGAUCUCUGAUCCAAUUG--------------CAAUUAGACUGGCGUGCUGCCCUCGGGACAGUUGUGGCAACAUCAAACCAUU (((.(((((((..((((((.(((((....))))).((((((...)))))).)))--------------)))......))))..((((((....)).))))..))).)))........... ( -32.40) >DroEre_CAF1 6171 119 + 1 UUUCCCAGCCAUUUUGCAUGGCAAAACAAUUUGCAUGGAUCUCCGAUCCAAUUGGCGAAAAAUUGGAAAAAUUAGACUGGCGUGCUGCCCUCGGGACAGCAGUGGCAACAUCG-CCCAUU (((((..(((((.....))))).......(((((.((((((...))))))....))))).....))))).........(((((((((((....)).)))))(((....)))))-)).... ( -39.70) >DroYak_CAF1 8871 93 + 1 GUUCCCAGCUAUGUUGCAUGGCAAAACAAUUUGCAUGGAUCUCUGAUCCAAUUG--------------CAAUUAGACUGGCGUGCUGCCCUCGGGAC------------AUUG-GCCAUU (((((.((....((.(((((.((.......(((((((((((...))))))..))--------------)))......)).))))).)).)).)))))------------....-...... ( -27.92) >DroAna_CAF1 10991 88 + 1 ----GUAGCAAUUUUACAUACCAAAGCAAUUUGCAUGGCUCUCCGAUCCACUUG--------------CAAUUAGGCUGGCGUGUUGCCCUCAGGAUGGCUGCAGC-------------- ----(((((.(((((.........(((...((((((((.((...)).)))..))--------------)))....)))(((.....)))...))))).)))))...-------------- ( -22.10) >consensus GUUCCCAGCCAUUUUGCAUGGCAAAACAAUUUGCAUGGAUCUCUGAUCCAAUUG______________CAAUUAGACUGGCGUGCUGCCCUCGGGACAGUUGUGGCAACAUCA_ACCAUU .......((((.((((....(((((....))))).((((((...))))))......................)))).))))..((((((....)).)))).................... (-18.02 = -18.72 + 0.70)

| Location | 19,192,895 – 19,193,000 |
|---|---|
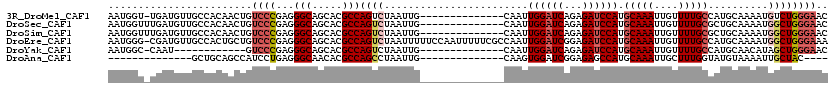
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19192895 105 - 27905053 AAUGGU-UGAUGUUGCCACAACUGUCCCGAGGGCAGCACGCCAGUCUAAUUG--------------CAAUUGGAUCAGAGAUCCAUGCAAAUUGUUUUGCCAUGCAAAAUGUCUGGGAAC ..((((-.......))))...((((((...))))))....((((.(...(((--------------((..((((((...)))))).(((((....)))))..)))))...).)))).... ( -31.50) >DroSec_CAF1 4885 106 - 1 AAUGGUUUGAUGUUGCCACAACUGUCCCGAGGGCAGCACGCCAGUCUAAUUG--------------CAAUUGGAUCAGAGAUCCAUGCAAAUUGUUUUGCGCUGCAAAAUGGCUGGGAAC ..((((........))))...((((((...))))))....((((((...(((--------------((..((((((...)))))).(((((....)))))..)))))...)))))).... ( -35.80) >DroSim_CAF1 5261 106 - 1 AAUGGUUUGAUGUUGCCACAACUGUCCCGAGGGCAGCACGCCAGUCUAAUUG--------------CAAUUGGAUCAGAGAUCCAUGCAAAUUGUUUUGCGCUGCAAAAUGGCUGGGAAC ..((((........))))...((((((...))))))....((((((...(((--------------((..((((((...)))))).(((((....)))))..)))))...)))))).... ( -35.80) >DroEre_CAF1 6171 119 - 1 AAUGGG-CGAUGUUGCCACUGCUGUCCCGAGGGCAGCACGCCAGUCUAAUUUUUCCAAUUUUUCGCCAAUUGGAUCGGAGAUCCAUGCAAAUUGUUUUGCCAUGCAAAAUGGCUGGGAAA ....((-((.(((((((.((.(......))))))))))))))........(((((((.......((....((((((...)))))).))..........(((((.....)))))))))))) ( -39.50) >DroYak_CAF1 8871 93 - 1 AAUGGC-CAAU------------GUCCCGAGGGCAGCACGCCAGUCUAAUUG--------------CAAUUGGAUCAGAGAUCCAUGCAAAUUGUUUUGCCAUGCAACAUAGCUGGGAAC ....((-...(------------((((...)))))))...((((.(((.(((--------------((..((((((...)))))).(((((....)))))..)))))..))))))).... ( -27.50) >DroAna_CAF1 10991 88 - 1 --------------GCUGCAGCCAUCCUGAGGGCAACACGCCAGCCUAAUUG--------------CAAGUGGAUCGGAGAGCCAUGCAAAUUGCUUUGGUAUGUAAAAUUGCUAC---- --------------...((((((........))).(((..((((..((((((--------------((..(((.((...)).)))))).)))))..))))..))).....)))...---- ( -21.80) >consensus AAUGGU_UGAUGUUGCCACAACUGUCCCGAGGGCAGCACGCCAGUCUAAUUG______________CAAUUGGAUCAGAGAUCCAUGCAAAUUGUUUUGCCAUGCAAAAUGGCUGGGAAC ........................((((...(((.....)))((((........................((((((...)))))).(((((....)))))..........)))))))).. (-17.99 = -18.93 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:40 2006