
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,192,543 – 19,192,750 |
| Length | 207 |
| Max. P | 0.957075 |

| Location | 19,192,543 – 19,192,639 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19192543 96 - 27905053 UUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGACUCCAUUGGGCCACUAUACGAUGGGUUAGGAGACUACCUUUGUUGGGAAAAUUAGAAA--------AAAA .((((.((((((((.....))))))))))))((.(((....))))).(((.((((.((((.((....)))))))))))))............--------.... ( -26.10) >DroEre_CAF1 5828 95 - 1 UUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGAU---GUU-GGCCACUAUACGAUGGGUUAAGAGUCUACCUUUGUGCGGAAAAUCA----AAUAAUUU-GUA ((((((((((((...))))))))(((((..(((.(---.((-((((.(........))))))).).....)))..))))).....)))----).......-... ( -22.30) >DroYak_CAF1 8506 95 - 1 UUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGAU---AUU-GGCCACUAUACGAUGGGUUAAGAGUCCACCUUUGUGCGGAAAAUAA----AAUAGUUU-UCA .((((.((((((((.....))))))))))))((((---.((-((((.(........)))))))..))))...........((((((..----....))))-)). ( -25.70) >consensus UUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGAU___AUU_GGCCACUAUACGAUGGGUUAAGAGUCUACCUUUGUGCGGAAAAUAA____AAUA_UUU_AAA (((((.((((((((.....)))))))))))))...............((.(((((.((((.........))))))))).))....................... (-18.00 = -18.33 + 0.33)


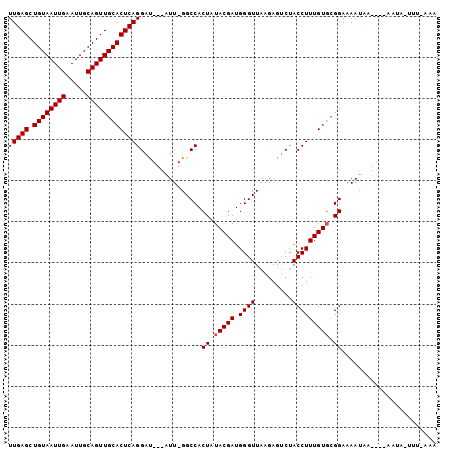
| Location | 19,192,572 – 19,192,670 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -27.80 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19192572 98 - 27905053 AUGGCCAUGGCUCUCUGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGACUCC--------AUUGGGCCAC-UAUACGAUGGGUUAGGAGA (((((..((((.....))))..))))).....(((((((((((...))))))))).))(((..(((.((--------((((......-....)))))))))..))). ( -34.50) >DroSec_CAF1 4557 107 - 1 AUGGCCAUGGCUCUCUGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGACUCCAAUGCACUAUUGGGCCACUUAUACGAUGGGUUAAAACA (((((..((((.....))))..))))).....((((.((((((((.....))))))))))))((.(.((((((...)))))))))((((((...))))))....... ( -31.70) >DroSim_CAF1 4919 107 - 1 AUGGCCAUGGCUCUCUGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGACUCCAAUGCACUAUUGGGCCACUUAUACGAUGGGUUAGGACA (((((..((((.....))))..)))))....(((((.((((((((.....)))))))))))))...(((...((.((((((...........))))))))..))).. ( -33.50) >DroEre_CAF1 5860 94 - 1 AUGGCCAUGGCUCUCUGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGAU-----------GUU-GGCCAC-UAUACGAUGGGUUAAGAGU (((((..((((.....))))..)))))....(((((.((((((((.....)))))))))))))..(-----------.((-((((.(-........))))))).).. ( -30.40) >DroYak_CAF1 8538 94 - 1 AUGGCCAUGGCUCUCUGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGAU-----------AUU-GGCCAC-UAUACGAUGGGUUAAGAGU (((((..((((.....))))..)))))....(((((.((((((((.....)))))))))))))...-----------.((-((((.(-........))))))).... ( -30.00) >consensus AUGGCCAUGGCUCUCUGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGUUGCACUCAGGACUCC________AUUGGGCCAC_UAUACGAUGGGUUAAGAGA .((((((((((...........)))))....(((((.((((((((.....)))))))))))))..................)))))..................... (-27.80 = -27.80 + 0.00)



| Location | 19,192,603 – 19,192,710 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -31.03 |
| Energy contribution | -30.76 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19192603 107 - 27905053 CGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGU----UGCACUCAGGACU ((((((((................))))))))........(((((..((((....---------.))))..)))))....(((((.((((((((.....))))----))))))))).... ( -36.29) >DroSec_CAF1 4597 107 - 1 CGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGU----UGCACUCAGGACU ((((((((................))))))))........(((((..((((....---------.))))..)))))....(((((.((((((((.....))))----))))))))).... ( -36.29) >DroSim_CAF1 4959 107 - 1 CGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGU----UGCACUCAGGACU ((((((((................))))))))........(((((..((((....---------.))))..)))))....(((((.((((((((.....))))----))))))))).... ( -36.29) >DroEre_CAF1 5888 106 - 1 CGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGU----UGCACUCAGGAU- ((((((((................))))))))........(((((..((((....---------.))))..)))))....(((((.((((((((.....))))----)))))))))...- ( -36.29) >DroYak_CAF1 8566 106 - 1 CGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGU----UGCACUCAGGAU- ((((((((................))))))))........(((((..((((....---------.))))..)))))....(((((.((((((((.....))))----)))))))))...- ( -36.29) >DroAna_CAF1 10666 119 - 1 CGCGGCGGUUCCAUAAACAAAUUGUCGCCGGGCUCUCUAAAUGGCCAUGGCUCUCAGUGGCUGCUGCUGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCACUGCACUGUACUCAGGAU- .((((((((..((.........))..))))((((........))))(((((((...(((((.((....)).)))))......)))))))............)))).(((....)))...- ( -35.60) >consensus CGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC_________UGCCGCUGCCAUGAAAUUGAGCUGUAAUUGAAUUGCAGU____UGCACUCAGGAC_ .(((((((((.........)))))))))...((((.....(((((..((((..............))))..)))))......)))).....((((..((((......)))).)))).... (-31.03 = -30.76 + -0.28)

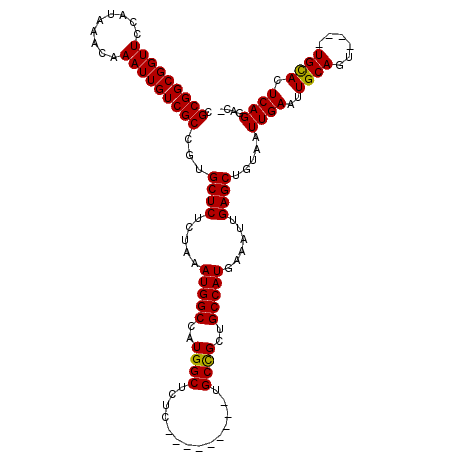

| Location | 19,192,639 – 19,192,750 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -33.14 |
| Energy contribution | -33.64 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19192639 111 - 27905053 GCCAAGAGCGAGUGAUUUAUGCACCGCACCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAA .......(((.(((.......)))))).(((((((.((..((((((((................))))))))..)).))))))).((((((....---------.......))))))... ( -36.69) >DroSec_CAF1 4633 111 - 1 GCCAAGAGCGAGUGAUUUAUGCACCGCACCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAA .......(((.(((.......)))))).(((((((.((..((((((((................))))))))..)).))))))).((((((....---------.......))))))... ( -36.69) >DroSim_CAF1 4995 111 - 1 GCCAAGAGCGAGUGAUUUAUGCACCGCACCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAA .......(((.(((.......)))))).(((((((.((..((((((((................))))))))..)).))))))).((((((....---------.......))))))... ( -36.69) >DroEre_CAF1 5923 111 - 1 GCCAGAAGCGAGUGAUUUAUGCACCGCACCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAA .......(((.(((.......)))))).(((((((.((..((((((((................))))))))..)).))))))).((((((....---------.......))))))... ( -36.69) >DroYak_CAF1 8601 111 - 1 GCCAAGAGCGAGUGAUUUAUGCACCGCACCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC---------UGCCGCUGCCAUGAAA .......(((.(((.......)))))).(((((((.((..((((((((................))))))))..)).))))))).((((((....---------.......))))))... ( -36.69) >DroAna_CAF1 10705 120 - 1 GCCGAGAGCGAGUGAUUUAUGCUCCGGUCCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGGGCUCUCUAAAUGGCCAUGGCUCUCAGUGGCUGCUGCUGCUGCCAUGAAA ...((((((((((.......)))).((.(((((((.((((((((((((((.........)))))))).))))))...)))))))))...)))))).(((((.((....)).))))).... ( -50.70) >consensus GCCAAGAGCGAGUGAUUUAUGCACCGCACCAUUUAAAGUCCGCGGCGGUUCCAUAAACAAAUUGUCGCCGUGCUCUCUAAAUGGCCAUGGCUCUC_________UGCCGCUGCCAUGAAA .......(((.(((.......)))))).(((((((.((..((((((((................))))))))..)).))))))).((((((....................))))))... (-33.14 = -33.64 + 0.50)


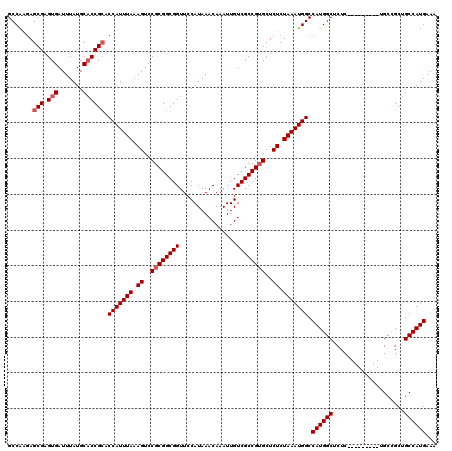
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:38 2006