
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,190,176 – 19,190,474 |
| Length | 298 |
| Max. P | 0.995008 |

| Location | 19,190,176 – 19,190,283 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19190176 107 - 27905053 UGGUUGGAUGGAUUGCAAUGGUAAAGCGGUUGCAUAAUCCCAUGUUAUGCAGUUUUGAAUUGAGCGAGAAAACUUCUUCAAAGCGAAGCGUGUUAAGGGUACAAACA ..(((((((....((((((.(.....).))))))..))))..(((...(((((((((..(((((.(((....))).)))))..)))))).))).......)))))). ( -25.70) >DroSec_CAF1 2321 96 - 1 UGGUUAAAUGGAUUGCAAUGGUAAAGCAGUUGCAUAAUGCCAUGUUAUGCAGUUGUGAAUUGAGCGAGAAAACUUCUUCAAAGCGAAGCGUGUAA-----------C ..((((.(((..((((.........(((..((((((((.....))))))))..)))...(((((.(((....))).))))).))))..))).)))-----------) ( -23.50) >DroSim_CAF1 2349 83 - 1 UGGUUGGAUGGAUUGCAAUGGUAAAGCAGUUGCAUAAUGCCAUGUUAUGCAGUUGUGAAUUGAGCGAGAAAACUUCUUCAA-------------A-----------C ..((((.(.....).))))......(((..((((((((.....))))))))..)))...(((((.(((....))).)))))-------------.-----------. ( -17.90) >DroEre_CAF1 3519 106 - 1 UGGCUGGAUGGAUUGCAAUGUUGAAACAAUUGCAUAAUGCCAUGUUGUGCAGUUGUAAAUUGAGUGAGAAAACUUCUUCGAAGCGAAGCGUGCUAAGGGUACAU-UG ..(((.......((((.........(((((((((((((.....)))))))))))))...(((((.(((....))).))))).)))))))((((.....))))..-.. ( -27.51) >DroYak_CAF1 6058 96 - 1 UGGUUGGAUGGAUUGCGAUGUUGGAGCAAUUGCAUAAUGCCAUGUUAUGCAGUUGCAAAUUGAGUGAGAAAACUUCUUCGAAGCGCAGCGUGUUAA----------- ......((((..(((((...((((((((((((((((((.....))))))))))))).....((((......)))).)))))..)))))..))))..----------- ( -29.30) >consensus UGGUUGGAUGGAUUGCAAUGGUAAAGCAGUUGCAUAAUGCCAUGUUAUGCAGUUGUGAAUUGAGCGAGAAAACUUCUUCAAAGCGAAGCGUGUUAA__________C ............((((.........(((((((((((((.....)))))))))))))...(((((.(((....))).))))).))))..................... (-17.96 = -18.04 + 0.08)



| Location | 19,190,216 – 19,190,323 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -22.56 |
| Consensus MFE | -20.26 |
| Energy contribution | -19.58 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19190216 107 - 27905053 AAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAUUGGUUGGAUGGAUUGCAAUGGUAAAGCGGUUGCAUAAUCCCAUGUUAUGCAGUUUUGAAUUGAGCGA ...((((((((((((((.....))))(((((..(((......)))((((....((((((.(.....).))))))..)))).....)))))..))))).))))).... ( -21.80) >DroSec_CAF1 2350 107 - 1 AAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAUUGGUUAAAUGGAUUGCAAUGGUAAAGCAGUUGCAUAAUGCCAUGUUAUGCAGUUGUGAAUUGAGCGA ...((((((........(((...(((((......)))))....)))......((((....)))).(((..((((((((.....))))))))..))).)))))).... ( -20.00) >DroSim_CAF1 2365 107 - 1 AAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAUUGGUUGGAUGGAUUGCAAUGGUAAAGCAGUUGCAUAAUGCCAUGUUAUGCAGUUGUGAAUUGAGCGA ....(((((..(((((.....)))))((......)).)))))(((.(((...((((....)))).(((..((((((((.....))))))))..)))..))).))).. ( -20.80) >DroEre_CAF1 3558 107 - 1 AAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAUUGGCUGGAUGGAUUGCAAUGUUGAAACAAUUGCAUAAUGCCAUGUUGUGCAGUUGUAAAUUGAGUGA .((((((((..(((((.....)))))((......)).)))))))).......(..(...(((...(((((((((((((.....))))))))))))).)))...)..) ( -24.70) >DroYak_CAF1 6087 107 - 1 AAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAUUGGUUGGAUGGAUUGCGAUGUUGGAGCAAUUGCAUAAUGCCAUGUUAUGCAGUUGCAAAUUGAGUGA .(.((((((..............(((((......)))))...((((.(.....).))))......(((((((((((((.....))))))))))))).)))))).).. ( -25.50) >consensus AAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAUUGGUUGGAUGGAUUGCAAUGGUAAAGCAGUUGCAUAAUGCCAUGUUAUGCAGUUGUGAAUUGAGCGA ...((((((..............(((((......)))))..........................(((((((((((((.....))))))))))))).)))))).... (-20.26 = -19.58 + -0.68)



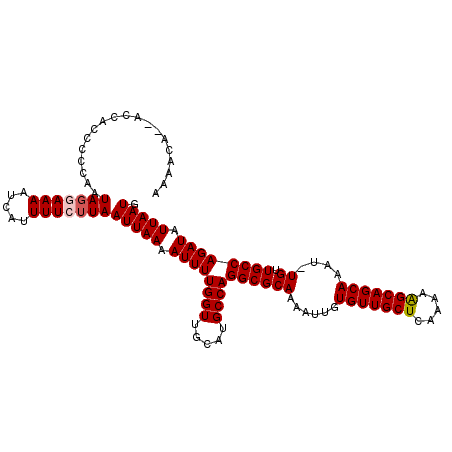
| Location | 19,190,283 – 19,190,402 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.15 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -27.27 |
| Energy contribution | -27.13 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19190283 119 - 27905053 GUUGCUCAAAAAGCAGCAAAUUUGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAU (((.(((.....((((((....))))))..(((.((((((((((((.(...((((......)-)))..).)))))))))))))))......))).))).....(((((......))))). ( -29.20) >DroSec_CAF1 2417 118 - 1 GUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAU (((.(((.....(((((....-.)))))..(((.((((((((((((.(...((((......)-)))..).)))))))))))))))......))).))).....(((((......))))). ( -28.60) >DroSim_CAF1 2432 118 - 1 GUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAU (((.(((.....(((((....-.)))))..(((.((((((((((((.(...((((......)-)))..).)))))))))))))))......))).))).....(((((......))))). ( -28.60) >DroEre_CAF1 3625 118 - 1 GUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAU (((.(((.....(((((....-.)))))..(((.((((((((((((.(...((((......)-)))..).)))))))))))))))......))).))).....(((((......))))). ( -28.60) >DroYak_CAF1 6154 119 - 1 GUUGCUCAAAAGGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUACUCCAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAU (((.(((....((((((....-.)))))).(((.((((((((((((.(...((((.......))))..).)))))))))))))))......))).))).....(((((......))))). ( -35.30) >DroAna_CAF1 8924 117 - 1 GUUGCUCAAAAGGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGAA- (((.(((....((((((....-.)))))).(((.((((((((((((.(...((((......)-)))..).)))))))))))))))......))).))).....(((((......)))))- ( -32.70) >consensus GUUGCUCAAAAAGCAGCAAAU_UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU_CAAGUCCAAGUCACGUUAAGUCAAUUAAGAGAAAUGAAUUUUCGUAUAUGAACGGAU (((.(((.....((((((....))))))..(((.((((((((((((.(...(((.........)))..).)))))))))))))))......))).))).....(((((......))))). (-27.27 = -27.13 + -0.14)



| Location | 19,190,323 – 19,190,442 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -28.86 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19190323 119 - 27905053 CUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAUUUGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUU ..............((((.....))))((((((((....(((((((.....)))))))..)))).)))).......((((((((((.(...((((......)-)))..).)))))))))) ( -32.10) >DroSec_CAF1 2457 118 - 1 CUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUU ..............((((.....))))(((((((.....(((((((.....)))))))..)-)).)))).......((((((((((.(...((((......)-)))..).)))))))))) ( -30.50) >DroSim_CAF1 2472 118 - 1 CUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUU ..............((((.....))))(((((((.....(((((((.....)))))))..)-)).)))).......((((((((((.(...((((......)-)))..).)))))))))) ( -30.50) >DroEre_CAF1 3665 118 - 1 CUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU-CAAGUCCAAGUCACGUU ..............((((.....))))(((((((.....(((((((.....)))))))..)-)).)))).......((((((((((.(...((((......)-)))..).)))))))))) ( -30.50) >DroYak_CAF1 6194 119 - 1 CUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAGGCAGCAAAU-UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUACUCCAAGUCCAAGUCACGUU ..............((((.....))))(((((((.....(((((((.....)))))))..)-)).)))).......((((((((((.(...((((.......))))..).)))))))))) ( -33.00) >consensus CUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU_UGUUGCCAGAUAUUAAUGUGGCUUUGUAAUUGGUUUUUCU_CAAGUCCAAGUCACGUU ..............((((.....))))((((((......(((((((.....)))))))....)).)))).......((((((((((.(...(((.........)))..).)))))))))) (-28.86 = -28.70 + -0.16)



| Location | 19,190,362 – 19,190,474 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19190362 112 - 27905053 AAAACA--ACCACCCCCAAUAGGAAAAUCAUUUUCUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAUUUGUUGCCAGAUAUUAAUG ....((--((((.......(((((((.....)))))))..........)))))).......((((((((....(((((((.....)))))))..)))).))))........... ( -25.13) >DroSec_CAF1 2496 108 - 1 ---ACA--ACCACCCCCAAUAGGAAAAUCAUUUUCUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUG ---.((--((((.......(((((((.....)))))))..........)))))).......(((((((.....(((((((.....)))))))..)-)).))))........... ( -23.53) >DroSim_CAF1 2511 108 - 1 ---ACA--ACCACCCCCAAUAGGAAAAUCAUUUUCUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUG ---.((--((((.......(((((((.....)))))))..........)))))).......(((((((.....(((((((.....)))))))..)-)).))))........... ( -23.53) >DroEre_CAF1 3704 110 - 1 AAAACA--AGCACCCCCAAUA-CAAAAUGAUUUUCUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU-UGUUGCCAGAUAUUAAUG ......--.(((...((((..-.....(((((.....))))).....)))).)))......(((((((.....(((((((.....)))))))..)-)).))))........... ( -20.50) >DroYak_CAF1 6234 113 - 1 AAAACACAGCCACCCCCAAUAGCAAAAUGAUUUUCUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAGGCAGCAAAU-UGUUGCCAGAUAUUAAUG ..(((((((............((((...((((((.......))))))....)))).(((.....)))...)))))))........((((((....-.))))))........... ( -22.90) >consensus AAAACA__ACCACCCCCAAUAGGAAAAUCAUUUUCUUAAUUAAAAUUUUGGUUGCAUGCCAGGCGCAAAAUUGUGUUGCUCAAAAAGCAGCAAAU_UGUUGCCAGAUAUUAAUG ...................(((((((.....)))))))(((((.((((((((.....))))((((((......(((((((.....)))))))....)).)))))))).))))). (-21.00 = -21.44 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:30 2006