
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,139,148 – 19,139,266 |
| Length | 118 |
| Max. P | 0.706220 |

| Location | 19,139,148 – 19,139,266 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -24.07 |
| Energy contribution | -24.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19139148 118 - 27905053 UUGCCACCCCUCACGCCGGGCACCAAUCGCAAGGUCAACGAGGUACUGAAGGCAUCCUUUGCCUCCUGGGAAAAGGAGGUCCAAAAAUGCAACAUAACCAAAGGUAAGAAAAU-GCGUA (((((...((((..(((..((.......))..)))....))))........((((.....(((((((......)))))))......))))............)))))......-..... ( -31.30) >DroVir_CAF1 22016 118 - 1 UUGCCGCCCCUCACGCCCGGCACAAACCGGAAGGUGAAUGAGGUUCUGAAAGCCUCAUUUGCCUCCUGGGAAAAGGAGGUGCAAAACUGUAAUAUAACCAAAGGUAAGAAAUA-AAAAA (((((((.((((...(((((......))(((.((..(((((((((.....)))))))))..))))).))).....)))).))......((......))....)))))......-..... ( -42.00) >DroGri_CAF1 22426 118 - 1 AUGCCACCCCUCACACCUGGCACGAAUCGCAAGGUGAACGAGGUGCUGAAAGCCUCGUUUGCCUCCUGGGAAAAGGAGGUGCAGAACUUUAAUAUAACCAAAGGUAAGGAGAG-CAACA .((((.(((((..(((((.((.......)).)))))((((((((.......)))))))).(((((((......))))))).....................)))...)).).)-))... ( -37.60) >DroWil_CAF1 27279 119 - 1 CUGCCACCCCUCACACCGGGCACCAAUCGCAAAGUCAACGAGGUGCUCAAAGCCUCAUUUGCCUCCUGGGAGAAGGAGGUGCAGAACUGCAACAUUACCAAAGGUAUGAAACUCAUUUU ..........(((.((((((((((..(((.........))))))))))............(((((((......)))))))(((....)))............))).))).......... ( -33.30) >DroMoj_CAF1 24036 118 - 1 UUGCCGCCCCUCACGCCUGGCACGAAUCGCAAGGUGAACGAGGUGCUGAAAGCCUCGUUUGCCUCCUGGGAGAAGGAGGUGCAGAACUGUAAUAUAACCAAAGGUAAGCAGCA-GCUCA .............(((((.((.......)).)))))...(((.(((((...((((.....(((((((......)))))))(((....)))...........))))...)))))-.))). ( -40.30) >DroAna_CAF1 19768 109 - 1 UUGCCACCCCUCACCCCAGGCACCAAUCGCAAGGUCAACGAGGUGCUCAAGGCCUCCUUUGCCUCCUGGGAGAAGGAGGUCCAAAACUGUAACAUAACCAAAGGUGGGU---------- ...(((((..........((((((.(((....)))......))))))...(((((((((..((.....))..))))))))).....................)))))..---------- ( -37.80) >consensus UUGCCACCCCUCACGCCGGGCACCAAUCGCAAGGUCAACGAGGUGCUGAAAGCCUCAUUUGCCUCCUGGGAAAAGGAGGUGCAAAACUGUAACAUAACCAAAGGUAAGAAAAA_AAACA .((((...((........))............(((....(((((.......)))))....(((((((......)))))))................)))...))))............. (-24.07 = -24.27 + 0.19)
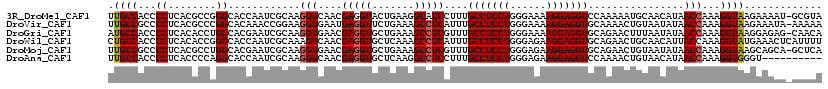
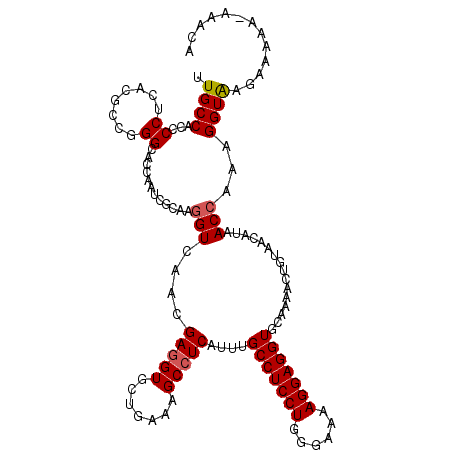
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:24 2006