
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,117,510 – 19,117,630 |
| Length | 120 |
| Max. P | 0.772733 |

| Location | 19,117,510 – 19,117,630 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -48.70 |
| Consensus MFE | -33.89 |
| Energy contribution | -35.08 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19117510 120 - 27905053 AUGAGCACCUUGGGUCUGGAUAAGGGCCUGCUGGGUGGCUAUACGACGCAGGGCGGAGUGCCGUGCUUCACGGGAUCGGGUCCCAUUCAACUGUGGCAAUUCCUGCUCGAAUUGCUGCUG .(((((((((.((((((......))))))...))))).......((.(((.(((.....))).))).))..(((((...))))).))))...(..(((((((......)))))))..).. ( -45.80) >DroVir_CAF1 435 120 - 1 AUGACCACAUUGGGUCUGGACAAAACGCUGCUGGGCGGCUACACAACGCAGGGCGGAGUGCCCUGUUUUACAGGCUCCGGGCCAAUACAACUGUGGCAAUUUCUGCUGGAAUUGCUGCUG ............((((((((.....((((....))))(((.......(((((((.....)))))))......))))))))))).........(..((((((((....))))))))..).. ( -45.52) >DroGri_CAF1 438 120 - 1 AUGACCACAUUGGGUCUGGAUAAAACGCUGCUAGGCGGCUACACAACGCAGGGGGGCGUGCCCUGCUUUACGGGCUCUGGUCCCAUUCAGUUGUGGCAAUUCCUGCUGGAGCUGUUGCUG ..(((.....((((.(..((....(((((.((..(((.........)))..)).)))))((((........))))))..).))))....)))(..(((.((((....)))).)))..).. ( -45.80) >DroWil_CAF1 522 111 - 1 ---------CUGGGUCUGGACAAAUCCCUACUGGGUGGCUAUACCACCCAAGGGGGAGUGCCCUGUUUCACGGGCUCCGGACCCAUACAGUUGUGGCAAUUCCUGCUCGAAUUAUUGCUG ---------.((((((((((....(((((..(((((((.....)))))))..)))))..((((........)))))))))))))).......(..(.(((((......))))).)..).. ( -51.30) >DroMoj_CAF1 443 120 - 1 AUGACCACAUUGGGUCUAGACAAAACGCUGCUGGGCGGCUAUACAACGCAGGGCGGUGUACCGUGUUUUACGGGCUCCGGACCCAUACAGCUGUGGCAGUUCCUGUUGGAGCUUCUGCUC .((.(((((.((((((..(((...((((((((..(((.........)))..)))))))).(((((...)))))).))..))))))......)))))))(((((....)))))........ ( -47.40) >DroAna_CAF1 417 120 - 1 AUGAGCACCCUGGGGCUGGACAAGGGCCUGCUAGGAGGCUACACGACGCAGGGCGGAGUGCCGUGCUUCACGGGCUCCGGUCCCAUCCAGCUGUGGCAGUUCCUGCUGGAGCUGCUCCUG ...(((....((((((((((.....(((((....(((((..(((..(((...)))..)))....))))).)))))))))))))))....)))(..((((((((....))))))))..).. ( -56.40) >consensus AUGACCACAUUGGGUCUGGACAAAACCCUGCUGGGCGGCUACACAACGCAGGGCGGAGUGCCCUGCUUCACGGGCUCCGGUCCCAUACAGCUGUGGCAAUUCCUGCUGGAACUGCUGCUG ..........((((((((((.......(((((..(((.........)))..)))))...(((.((.....))))))))))))))).......(..((((((((....))))))))..).. (-33.89 = -35.08 + 1.20)

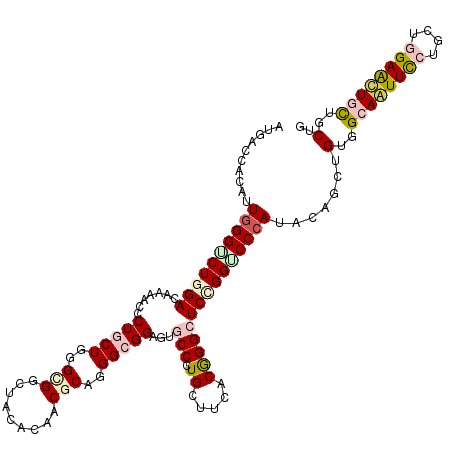
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:16 2006