
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,091,871 – 19,091,976 |
| Length | 105 |
| Max. P | 0.936820 |

| Location | 19,091,871 – 19,091,976 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19091871 105 + 27905053 GUUGCGAGUCAAGGGCUUGUGAUUUUUGACGAUGUCACAGCUGUGCUGCCAUUGCCAGCAGCUGCGGCAGAAGUAUCUGUAAACAGAAUGUAGGA---A-AUAAAAUAU- .(..(((((.....)))))..)((((..(((.((((.(((((((...((....))..))))))).))))......((((....)))).)))..))---)-)........- ( -31.60) >DroVir_CAF1 7922 103 + 1 GUUACGUGUCAAUGGCUUGUGAUUUUUAAUAAUUUCACAGCUGUGUUGCCAUUGCCAACAGCUGCGGCAAAAGUAACUGCAAC-AGA-UACAUGU---A-AUUUAGU-UA (((((((((...(((((.((((..((....))..)))))))((((((((..(((((.........)))))..))))).))).)-)..-.))))))---)-)).....-.. ( -28.80) >DroGri_CAF1 7614 104 + 1 GCUACGUGUCAAGGGCUUGUGAUUUUUAAUAAUUUCACAGCUGUGCUGCCAUUGCCAACAGCUGCGGCAAAAGUAACUGCAAC-ACAAUUAGUUU---C-AAUUAGU-UG .....((((..((((((.((((..((....))..)))))))).((((((..(((....)))..)))))).......))...))-))(((((((..---.-.))))))-). ( -24.60) >DroEre_CAF1 7850 106 + 1 GUUGCGCGUCAAGGGCUUGUGAUUUUUGACGAUGUCGCAGCUGUGCUGCCAUUGCCAGCAGCUGCGGCAGAAGUAUCUGCAAAUGGAAUAUAUGA---AAGUAAAGUAU- .((((.((((((((((....).))))))))).((((((((((((...((....))..)))))))))))).........))))........(((..---..)))......- ( -36.00) >DroMoj_CAF1 7612 103 + 1 AUUUCUUGUCAAAGGCUUGUGAUUUUUGAUAAUUUCACAGCUGUGCUGCCAUUGCCAACAGCUGCGGCAAAAGUAACUGCAAU-GGA-UAUGCAU---A-GGUAAAUAU- .....(((((((((((....).))))))))))........((((((..(((((((.....(((........)))....)))))-)).-...))))---)-)........- ( -26.00) >DroAna_CAF1 7509 106 + 1 GUUACGGGUCAAGGGUUUGUGAUUCUUGACGAUGUCACAGCUAUGCUGCCAUUGCCAGCAGCUGCGACAGAAGUAUCUGGAA-UAGAA-GUAGCUUUCC-UUUAAAUAU- (((((..((((((((((...))))))))))..((((.(((((..((((.......))))))))).)))).............-.....-))))).....-.........- ( -31.50) >consensus GUUACGUGUCAAGGGCUUGUGAUUUUUGACAAUGUCACAGCUGUGCUGCCAUUGCCAACAGCUGCGGCAAAAGUAACUGCAAA_AGAAUAUAUGU___A_AUUAAAUAU_ ..(((........((((.((((..((....))..)))))))).((((((..(((....)))..))))))...)))................................... (-16.64 = -16.33 + -0.30)



| Location | 19,091,871 – 19,091,976 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19091871 105 - 27905053 -AUAUUUUAU-U---UCCUACAUUCUGUUUACAGAUACUUCUGCCGCAGCUGCUGGCAAUGGCAGCACAGCUGUGACAUCGUCAAAAAUCACAAGCCCUUGACUCGCAAC -.........-.---........((((....))))......((((((((((((((.(...).))))..))))))).....(((((.............)))))..))).. ( -24.12) >DroVir_CAF1 7922 103 - 1 UA-ACUAAAU-U---ACAUGUA-UCU-GUUGCAGUUACUUUUGCCGCAGCUGUUGGCAAUGGCAACACAGCUGUGAAAUUAUUAAAAAUCACAAGCCAUUGACACGUAAC ..-......(-(---((.(((.-...-(((((((......))))(((((((((..((....))...)))))))))..................))).....))).)))). ( -23.60) >DroGri_CAF1 7614 104 - 1 CA-ACUAAUU-G---AAACUAAUUGU-GUUGCAGUUACUUUUGCCGCAGCUGUUGGCAAUGGCAGCACAGCUGUGAAAUUAUUAAAAAUCACAAGCCCUUGACACGUAGC ..-.......-.---...(((...((-(((((((......))))(((((((((..((....))...))))))))).........................))))).))). ( -24.80) >DroEre_CAF1 7850 106 - 1 -AUACUUUACUU---UCAUAUAUUCCAUUUGCAGAUACUUCUGCCGCAGCUGCUGGCAAUGGCAGCACAGCUGCGACAUCGUCAAAAAUCACAAGCCCUUGACGCGCAAC -...........---...............(((((....)))))(((((((((((.(...).))))..)))))))....((((((.............))))))...... ( -28.12) >DroMoj_CAF1 7612 103 - 1 -AUAUUUACC-U---AUGCAUA-UCC-AUUGCAGUUACUUUUGCCGCAGCUGUUGGCAAUGGCAGCACAGCUGUGAAAUUAUCAAAAAUCACAAGCCUUUGACAAGAAAU -.........-.---.((((..-...-..)))).....(((((.(((((((((..((....))...)))))))))......(((((...........))))))))))... ( -22.10) >DroAna_CAF1 7509 106 - 1 -AUAUUUAAA-GGAAAGCUAC-UUCUA-UUCCAGAUACUUCUGUCGCAGCUGCUGGCAAUGGCAGCAUAGCUGUGACAUCGUCAAGAAUCACAAACCCUUGACCCGUAAC -.........-((((((....-..)).-))))...(((...((((((((((((((.(...).))))..))))))))))..((((((...........))))))..))).. ( -30.70) >consensus _AUACUUAAU_U___ACAUAUAUUCU_GUUGCAGAUACUUCUGCCGCAGCUGCUGGCAAUGGCAGCACAGCUGUGAAAUCAUCAAAAAUCACAAGCCCUUGACACGUAAC ..............................((((......))))(((((((((((.(...).))))..)))))))................................... (-17.51 = -17.07 + -0.44)

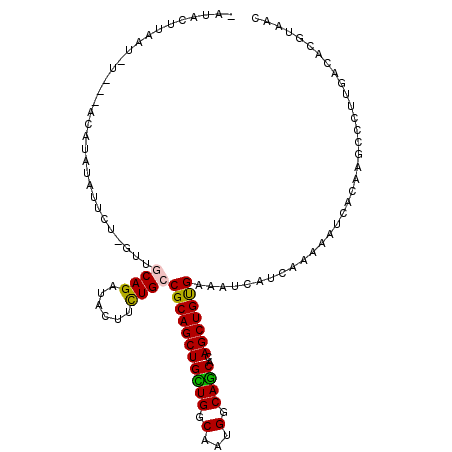
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:09 2006