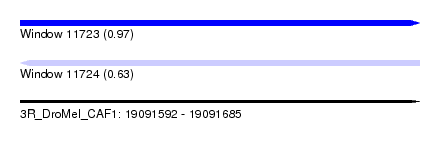
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,091,592 – 19,091,685 |
| Length | 93 |
| Max. P | 0.966381 |

| Location | 19,091,592 – 19,091,685 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 70.97 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -20.95 |
| Energy contribution | -19.57 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
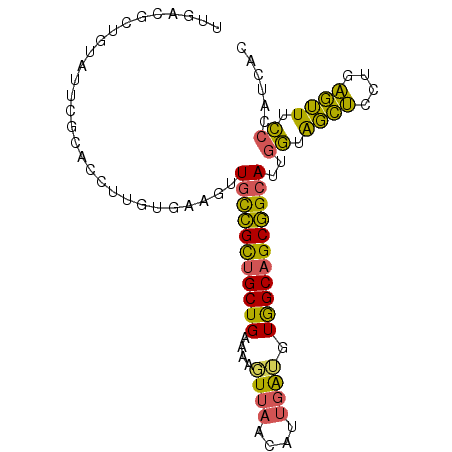
>3R_DroMel_CAF1 19091592 93 + 27905053 UUGACGCUGUAUUCGCACCUUGUGAAUUUGCCGCUGCUGAAGAGUUACCAUUGAUGUAGCAGCGGCAUUGGCAGCUCCUGAGUUUCCCAUCAC .....(((((((((((.....)))))).(((((((((((....((((....)))).)))))))))))...)))))...(((........))). ( -33.40) >DroSec_CAF1 7496 93 + 1 UUGACGCUGUAUUCGCACCUUUUGAAGUUGCCGCUGCUGAAGUGUUAACAUUGAGGUGGCAGCGGCAUUGGUAGCUCCUGAGUUUCCCAUCAC .....((.......))......(((...(((((((((((.((((....))))....))))))))))).(((.((((....))))..)))))). ( -27.80) >DroSim_CAF1 7432 93 + 1 UUGACGCUGUAUUCGCACCUUUUGAAGUUGCCGCUGCUGAAGAGUUAACAUUGAUGUGGCAGCGGCAUUGGUAGCUCCUGAGUUUCCCAUCAC .....((.......))......(((...(((((((((((....((((....)))).))))))))))).(((.((((....))))..)))))). ( -25.80) >DroEre_CAF1 7610 90 + 1 AUGAUGCUGCUGCCGCUCCUUCAGAAGUUGCCGCUGCUGAAAAGUUAACGUUGAUGUGGCAGCGGCAUUGGUGGCUCCCGAGUUUCCU---GG ..((((((((((((((...(((((.(((....))).)))))..((((....))))))))))))))))))((..(((....)))..)).---.. ( -36.00) >DroYak_CAF1 7341 93 + 1 CUGCUGCUGCUGCCGCUCCUUGAUAAGUUGUCGCUGCUGAAAAGUUAACGUUGGUGUGGCAGCGGCAUUGGUAGCUCCUGAGUUUCCCAUCUG ....((((((((((((.((..(((.....)))...(((....))).......)).)))))))))))).(((.((((....))))..))).... ( -33.70) >DroMoj_CAF1 7273 72 + 1 UUG---------------------GUGUUGCUGUUGCUGAUAUUUGUACAAUGAAUUGGCUGCAGAAUUGUUGCUGCCUGCAGUUGACAAGAG ...---------------------(((((((....)).)))))((((.((((.....(((.((((.....)))).)))....))))))))... ( -16.50) >consensus UUGACGCUGUAUUCGCACCUUGUGAAGUUGCCGCUGCUGAAAAGUUAACAUUGAUGUGGCAGCGGCAUUGGUAGCUCCUGAGUUUCCCAUCAC ............................(((((((((((....((((....)))).)))))))))))..((.((((....)))).))...... (-20.95 = -19.57 + -1.38)


| Location | 19,091,592 – 19,091,685 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 70.97 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -14.16 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19091592 93 - 27905053 GUGAUGGGAAACUCAGGAGCUGCCAAUGCCGCUGCUACAUCAAUGGUAACUCUUCAGCAGCGGCAAAUUCACAAGGUGCGAAUACAGCGUCAA .((((((....)).....((((....((((((((((...................)))))))))).((((.(.....).)))).)))))))). ( -29.31) >DroSec_CAF1 7496 93 - 1 GUGAUGGGAAACUCAGGAGCUACCAAUGCCGCUGCCACCUCAAUGUUAACACUUCAGCAGCGGCAACUUCAAAAGGUGCGAAUACAGCGUCAA .((((((....)......((.(((..(((((((((.....................))))))))).........)))))........))))). ( -24.40) >DroSim_CAF1 7432 93 - 1 GUGAUGGGAAACUCAGGAGCUACCAAUGCCGCUGCCACAUCAAUGUUAACUCUUCAGCAGCGGCAACUUCAAAAGGUGCGAAUACAGCGUCAA .((((((....)......((.(((..(((((((((.....................))))))))).........)))))........))))). ( -24.40) >DroEre_CAF1 7610 90 - 1 CC---AGGAAACUCGGGAGCCACCAAUGCCGCUGCCACAUCAACGUUAACUUUUCAGCAGCGGCAACUUCUGAAGGAGCGGCAGCAGCAUCAU ((---((....)).))..(((.....(((((((((.....................))))))))).((((....)))).)))........... ( -29.20) >DroYak_CAF1 7341 93 - 1 CAGAUGGGAAACUCAGGAGCUACCAAUGCCGCUGCCACACCAACGUUAACUUUUCAGCAGCGACAACUUAUCAAGGAGCGGCAGCAGCAGCAG ....(((....).))((.....))..(((.((((((........(((........))).((..(..........)..)))))))).))).... ( -21.90) >DroMoj_CAF1 7273 72 - 1 CUCUUGUCAACUGCAGGCAGCAACAAUUCUGCAGCCAAUUCAUUGUACAAAUAUCAGCAACAGCAACAC---------------------CAA ...........(((.(((.(((.......))).)))......((((..........))))..)))....---------------------... ( -10.60) >consensus CUGAUGGGAAACUCAGGAGCUACCAAUGCCGCUGCCACAUCAAUGUUAACUCUUCAGCAGCGGCAACUUCAAAAGGUGCGAAUACAGCGUCAA ....(((....((....))...))).(((((((((.....................))))))))).........(..((.......))..).. (-14.16 = -14.30 + 0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:07 2006