
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,077,222 – 19,077,313 |
| Length | 91 |
| Max. P | 0.924314 |

| Location | 19,077,222 – 19,077,313 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.885076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19077222 91 + 27905053 CCUACUGUGACGUAGAGCAACUCUGGCCGACAGCUG-UUUGGCCAGUGUUGACAAGCGCCACGCCGUCAAAAGCAAAUCUCAUUUAGUUUCC ......(((.(((....((((.(((((((((....)-.)))))))).))))....))).)))((........)).................. ( -26.40) >DroSec_CAF1 91487 91 + 1 CCUACUGUGACGUAGAACAACUCUGGCCGACAGCUG-UUUGGCCAGUGUUGACAAACGCCACGCCGUCAAAAGCAAAUCUCAUUUAGUUUCC ......(((.(((....((((.(((((((((....)-.)))))))).))))....))).)))((........)).................. ( -25.00) >DroSim_CAF1 94571 91 + 1 CCUACUGUGACGUAGGGCAACUCUGGCCGACAGCUG-UUCGGCCAGUGUUGACAAACGCCACGCAGUCAAAAGCAAAUCUCAUUUAGUUUCC ...((((((.(((..(.((((.(((((((((....)-.)))))))).)))).)..)))...))))))......................... ( -29.70) >DroEre_CAF1 102800 92 + 1 CCUGCUGUGACGUAGGUCAACUCUGGCCGACAGCUGGUUCAGCCAGUGUUGACAAACGCCACGCCGUCAAAAGCAAAUCUCAUUUAGUUUCC ..((((.((((((..((((((.(((((.((........)).))))).))))))..))((...)).))))..))))................. ( -28.50) >DroYak_CAF1 97420 92 + 1 CCUACUGGCACGUAGAGCAACUCUGGCCGACAGCUGGUUGAGCCAGUGUUGACAAACGCCACGCCGUCAAAAGCAAAUCUCAUUUAGUUUCC ......(((.(((....((((.(((((((((.....)))).))))).))))....)))....)))........................... ( -24.40) >consensus CCUACUGUGACGUAGAGCAACUCUGGCCGACAGCUG_UUCGGCCAGUGUUGACAAACGCCACGCCGUCAAAAGCAAAUCUCAUUUAGUUUCC ...((((((.(((....((((.((((((((........)))))))).))))....))).)))((........))...........))).... (-22.76 = -23.28 + 0.52)



| Location | 19,077,222 – 19,077,313 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19077222 91 - 27905053 GGAAACUAAAUGAGAUUUGCUUUUGACGGCGUGGCGCUUGUCAACACUGGCCAAA-CAGCUGUCGGCCAGAGUUGCUCUACGUCACAGUAGG .....(((..........(((......)))((((((...(.((((.((((((..(-(....)).)))))).)))))....))))))..))). ( -27.60) >DroSec_CAF1 91487 91 - 1 GGAAACUAAAUGAGAUUUGCUUUUGACGGCGUGGCGUUUGUCAACACUGGCCAAA-CAGCUGUCGGCCAGAGUUGUUCUACGUCACAGUAGG .....(((..........(((......)))(((((((....((((.((((((..(-(....)).)))))).))))....)))))))..))). ( -30.60) >DroSim_CAF1 94571 91 - 1 GGAAACUAAAUGAGAUUUGCUUUUGACUGCGUGGCGUUUGUCAACACUGGCCGAA-CAGCUGUCGGCCAGAGUUGCCCUACGUCACAGUAGG (....)....................(((((((((((..(.((((.((((((((.-(....))))))))).)))).)..))))))).)))). ( -36.70) >DroEre_CAF1 102800 92 - 1 GGAAACUAAAUGAGAUUUGCUUUUGACGGCGUGGCGUUUGUCAACACUGGCUGAACCAGCUGUCGGCCAGAGUUGACCUACGUCACAGCAGG (....)............(((......)))(((((((..((((((.((((((((........)))))))).))))))..)))))))...... ( -37.50) >DroYak_CAF1 97420 92 - 1 GGAAACUAAAUGAGAUUUGCUUUUGACGGCGUGGCGUUUGUCAACACUGGCUCAACCAGCUGUCGGCCAGAGUUGCUCUACGUGCCAGUAGG (....)..........(((((...((((((.(((.....((((....))))....)))))))))((((((((...))))..).)))))))). ( -24.50) >consensus GGAAACUAAAUGAGAUUUGCUUUUGACGGCGUGGCGUUUGUCAACACUGGCCAAA_CAGCUGUCGGCCAGAGUUGCUCUACGUCACAGUAGG (....)............((........))(((((((....((((.((((((............)))))).))))....)))))))...... (-26.16 = -26.52 + 0.36)

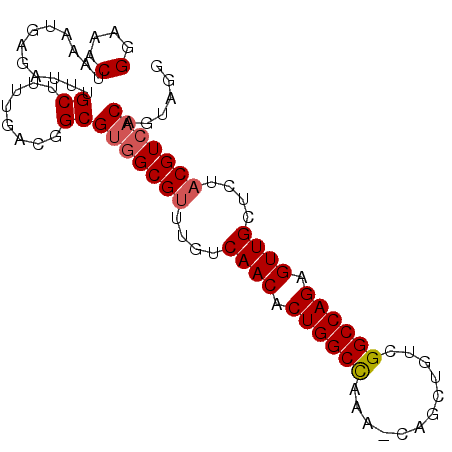
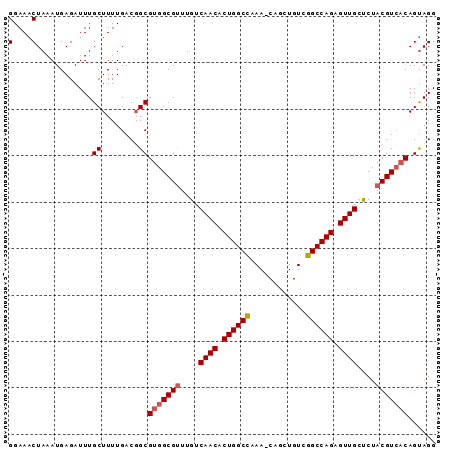
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:02 2006