
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,068,454 – 19,068,545 |
| Length | 91 |
| Max. P | 0.994314 |

| Location | 19,068,454 – 19,068,545 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -22.61 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19068454 91 + 27905053 AGAGUUGGGCCCAAGUGCU-----CGAUAAGUGGGUCUU-GGGAUUGGCAUGUAGGAUUACCGCUGCCAUCGAU------UUCAAUGGCAUUGGGUUCAACUG ..((((((((((((.((((-----.(((......)))((-(..(((((((.((.((....)))))))))...))------..))).)))))))))))))))). ( -34.30) >DroSec_CAF1 82888 91 + 1 AGAGUUGGGCCCAAGUGCU-----CGAUAAGUGGGUCUU-GGGAUUGGCAUGUGGGGUUACCGCUGCCAUCGAU------UUCAAUGGCAUUGGGUUCAACUG ..((((((((((((.((((-----.(((......)))((-(..(((((((.((((.....)))))))))...))------..))).)))))))))))))))). ( -39.40) >DroSim_CAF1 85316 91 + 1 AGAGUUGGGCCCAAGUGCU-----CGAUAAGUGGGUCUU-GGGAUUGGCAUGUGGGGUUACCGCUGCCAUCGAU------UUCAAUGGCAUUGGGUUCAACUG ..((((((((((((.((((-----.(((......)))((-(..(((((((.((((.....)))))))))...))------..))).)))))))))))))))). ( -39.40) >DroEre_CAF1 92503 82 + 1 AGAGUUGGGCCCAAGUGCUCGGCUCGCUAAGUGGGUCUU-GGGAUGGGCAU--------------GCCAUCGAU------UUGAAUGGCAUUGGGUUCAACUG ..(((((((((((((((((((..((.(((((.....)))-)))))))))))--------------(((((....------....))))).)))))))))))). ( -36.70) >DroYak_CAF1 87706 95 + 1 AGAGUUGCGCCCAAGUGCUGGACUCGUUAAGUGGGUCUU-GGGAUUGGCAUGUG-GUUUAGCGCUGCCAUCGAU------UUGAUUGGCAUUGGGCUCAACUG ..(((((.(((((((((((((((((((...((.((((..-..)))).))))).)-)))))))))(((((((...------..)).))))))))))).))))). ( -41.30) >DroPer_CAF1 88183 93 + 1 AAAGUUCAGCUCGAGUGGU-----UAACCAGAGAGGGUUAAGGCUGUGCAUAGGG-GUUA----AGCAAUCGAUACCAGUAUGAAUGGCAUUGGGAUCAACUC ..((((...(((((....(-----(((((......)))))).(((((.((((..(-((..----..........)))..)))).))))).)))))...)))). ( -21.90) >consensus AGAGUUGGGCCCAAGUGCU_____CGAUAAGUGGGUCUU_GGGAUUGGCAUGUGGGGUUACCGCUGCCAUCGAU______UUCAAUGGCAUUGGGUUCAACUG ..((((((((((((................((.((((.....)))).))................(((((..............))))).)))))))))))). (-22.61 = -23.34 + 0.73)



| Location | 19,068,454 – 19,068,545 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -12.32 |
| Energy contribution | -13.54 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19068454 91 - 27905053 CAGUUGAACCCAAUGCCAUUGAA------AUCGAUGGCAGCGGUAAUCCUACAUGCCAAUCCC-AAGACCCACUUAUCG-----AGCACUUGGGCCCAACUCU .(((((..(((((((((((((..------..))))))))((((((........))))......-(((.....)))....-----.))..)))))..))))).. ( -28.40) >DroSec_CAF1 82888 91 - 1 CAGUUGAACCCAAUGCCAUUGAA------AUCGAUGGCAGCGGUAACCCCACAUGCCAAUCCC-AAGACCCACUUAUCG-----AGCACUUGGGCCCAACUCU .(((((..(((((((((((((..------..))))))))((((((........))))......-(((.....)))....-----.))..)))))..))))).. ( -28.40) >DroSim_CAF1 85316 91 - 1 CAGUUGAACCCAAUGCCAUUGAA------AUCGAUGGCAGCGGUAACCCCACAUGCCAAUCCC-AAGACCCACUUAUCG-----AGCACUUGGGCCCAACUCU .(((((..(((((((((((((..------..))))))))((((((........))))......-(((.....)))....-----.))..)))))..))))).. ( -28.40) >DroEre_CAF1 92503 82 - 1 CAGUUGAACCCAAUGCCAUUCAA------AUCGAUGGC--------------AUGCCCAUCCC-AAGACCCACUUAGCGAGCCGAGCACUUGGGCCCAACUCU .(((((......((((((((...------...))))))--------------))(((((....-(((.....))).((.......))...))))).))))).. ( -22.80) >DroYak_CAF1 87706 95 - 1 CAGUUGAGCCCAAUGCCAAUCAA------AUCGAUGGCAGCGCUAAAC-CACAUGCCAAUCCC-AAGACCCACUUAACGAGUCCAGCACUUGGGCGCAACUCU .(((((.(((((((((((.((..------...)))))))((((.....-.....)).......-..(((.(.......).)))..))..)))))).))))).. ( -26.50) >DroPer_CAF1 88183 93 - 1 GAGUUGAUCCCAAUGCCAUUCAUACUGGUAUCGAUUGCU----UAAC-CCCUAUGCACAGCCUUAACCCUCUCUGGUUA-----ACCACUCGAGCUGAACUUU (((((((((...((((((.......)))))).))))((.----....-......)).(((((((((((......)))))-----)......).))))))))). ( -16.20) >consensus CAGUUGAACCCAAUGCCAUUCAA______AUCGAUGGCAGCGGUAACCCCACAUGCCAAUCCC_AAGACCCACUUAUCG_____AGCACUUGGGCCCAACUCU .(((((..((((((((................(((((((..............)))).)))...(((.....)))..........))).)))))..))))).. (-12.32 = -13.54 + 1.22)


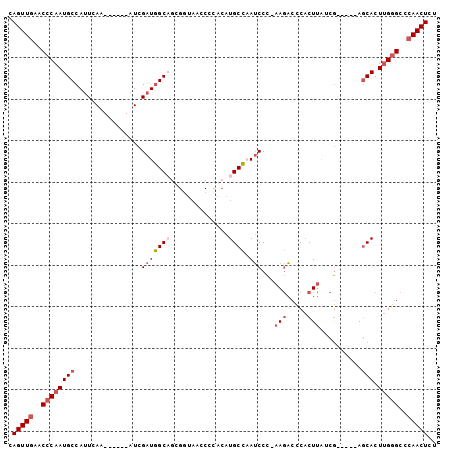
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:56 2006