
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,041,444 – 19,041,610 |
| Length | 166 |
| Max. P | 0.996840 |

| Location | 19,041,444 – 19,041,548 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -20.88 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19041444 104 + 27905053 AGCUUCAUUU-----------UUUUG---CUGCGCCAGUCUGCGUGUUGAGGUCACACAUA--UGUACGCAAUGUGGCGACCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC .((..((...-----------...))---..))(((.((((..(((((((((((.(((((.--((....)))))))..))))))((((....)))).)))))..))))..)))....... ( -34.10) >DroSec_CAF1 54056 107 + 1 AGCUUCAUUU-----------UUUUGCUGCUGCGCCAGUCUGCGUGUUGAGGUCACACAUA--UGUACGCAAUGUGGCGACCUCUCUCACCUGAGAGAACAUAAAGACCAGGUAGAAAUC (((..((...-----------...))..)))..(((.((((..(((((((((((.(((((.--((....)))))))..))))))((((....)))).)))))..))))..)))....... ( -34.80) >DroSim_CAF1 55751 107 + 1 AGCUUCAUUU-----------UUUUGCAGCUGCGACAGUCUGCGUGUUGAGGUCACACACA--UGUACGCAAUGUGGCGACCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC ((((.((...-----------...)).))))...((.((((..(((((((((((.(.((((--(.......)))))).))))))((((....)))).)))))..))))...))....... ( -33.00) >DroEre_CAF1 57009 102 + 1 AGCUUCAUUUUUGUU--UGUUUUUUGCU---GCGCCAGUCUGCGUGUUGAGGUCACACACA--UGUACGCAUUGUGGCGACCUCACUCACCUGAGAGAACAGAAAAACC----------- .......((((((((--(..........---((((......))))..(((((((.((((.(--((....)))))))..)))))))(((....))).)))))))))....----------- ( -30.10) >DroYak_CAF1 57377 120 + 1 AGCUUCAUUUCUGAUUUUUUUUUUUGCUUUUUUGCCAGUCUGCGUGUUGAGGUCACACACAUAUGUACGCAAUGUGGCGACCUCACUCACCCGAGAGAACAGUAAAACCAGGUAGACAUC .........................((......))..((((((.((.(((((((.(.(((((.((....)))))))).)))))))(((......)))...........)).))))))... ( -32.40) >consensus AGCUUCAUUU___________UUUUGCU_CUGCGCCAGUCUGCGUGUUGAGGUCACACACA__UGUACGCAAUGUGGCGACCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC .....................................((((..(((((((((((.((((....((....)).))))..)))))).(((......))))))))..))))............ (-20.88 = -21.68 + 0.80)


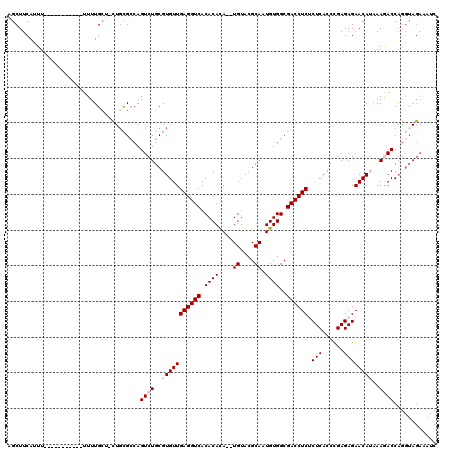
| Location | 19,041,444 – 19,041,548 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19041444 104 - 27905053 GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGGUCGCCACAUUGCGUACA--UAUGUGUGACCUCAACACGCAGACUGGCGCAG---CAAAA-----------AAAUGAAGCU .........(..((((.(.(((((((......))))((((((((.((((((....))--.)))))))))))).))).).))))..).((..---((...-----------...))..)). ( -29.90) >DroSec_CAF1 54056 107 - 1 GAUUUCUACCUGGUCUUUAUGUUCUCUCAGGUGAGAGAGGUCGCCACAUUGCGUACA--UAUGUGUGACCUCAACACGCAGACUGGCGCAGCAGCAAAA-----------AAAUGAAGCU ..((((...(..((((.(.((((.((((....))))((((((((.((((((....))--.)))))))))))))))).).))))..).((....))....-----------....)))).. ( -30.90) >DroSim_CAF1 55751 107 - 1 GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGGUCGCCACAUUGCGUACA--UGUGUGUGACCUCAACACGCAGACUGUCGCAGCUGCAAAA-----------AAAUGAAGCU ...........((.((((((..((((......))))(((((((((((((.......)--)))).)))))))).....((((.(((...)))))))....-----------..)))))))) ( -34.40) >DroEre_CAF1 57009 102 - 1 -----------GGUUUUUCUGUUCUCUCAGGUGAGUGAGGUCGCCACAAUGCGUACA--UGUGUGUGACCUCAACACGCAGACUGGCGC---AGCAAAAAACA--AACAAAAAUGAAGCU -----------.((((((((((.((.....(((..(((((((((((((.((....))--)))).)))))))))...))).....)).))---))..)))))).--............... ( -31.00) >DroYak_CAF1 57377 120 - 1 GAUGUCUACCUGGUUUUACUGUUCUCUCGGGUGAGUGAGGUCGCCACAUUGCGUACAUAUGUGUGUGACCUCAACACGCAGACUGGCAAAAAAGCAAAAAAAAAAAUCAGAAAUGAAGCU .........((((((((.......(((.(.(((..((((((((((((((((....)).))))).))))))))).))).))))...((......)).......)))))))).......... ( -34.90) >consensus GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGGUCGCCACAUUGCGUACA__UGUGUGUGACCUCAACACGCAGACUGGCGCAG_AGCAAAA___________AAAUGAAGCU .........(((((((...(((((((......))).(((((((((((..((....))...))).))))))))))))...))))))).................................. (-23.86 = -24.62 + 0.76)

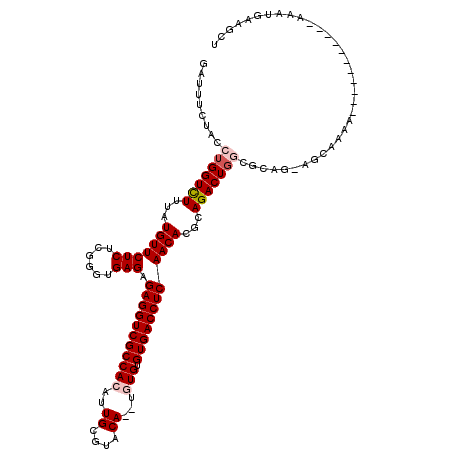

| Location | 19,041,470 – 19,041,577 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19041470 107 + 27905053 UGCGUGUUGAGGUCACACAUA--UGUACGCAAUGUGGCGACCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC---UACCUGAUCUCUCCGGAGAGAGA---AUGUGA .((((.....((((.(((((.--((....)))))))..))))((((((.((.((((((.(.....)..(((((((....)---)))))).)))))).)).))))))---)))).. ( -43.80) >DroSec_CAF1 54085 102 + 1 UGCGUGUUGAGGUCACACAUA--UGUACGCAAUGUGGCGACCUCUCUCACCUGAGAGAACAUAAAGACCAGGUAGAAAUC---UAUCUGAUCUCUGCGGAGA--------GGUGA ...(((((((((((.(((((.--((....)))))))..))))))((((....)))).)))))......(((((((....)---))))))((((((....)))--------))).. ( -36.50) >DroSim_CAF1 55780 105 + 1 UGCGUGUUGAGGUCACACACA--UGUACGCAAUGUGGCGACCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC---UACCUGAUCUCUGCGGAGA--GA---AGGUGA ........((((((.(.((((--(.......)))))).))))))((((..((((((((.(.....)..(((((((....)---)))))).))))).))).))--))---...... ( -39.40) >DroEre_CAF1 57044 89 + 1 UGCGUGUUGAGGUCACACACA--UGUACGCAUUGUGGCGACCUCACUCACCUGAGAGAACAGAAAAACC-------------------GAUCUCUGCGGAGA--GA---AGGUGA ...(((.(((((((.((((.(--((....)))))))..)))))))..)))(((......)))....(((-------------------..(((((....)))--))---.))).. ( -30.10) >DroYak_CAF1 57417 113 + 1 UGCGUGUUGAGGUCACACACAUAUGUACGCAAUGUGGCGACCUCACUCACCCGAGAGAACAGUAAAACCAGGUAGACAUCAACUAGCUGAUCUCUGCGGAGA--GAAAAAGGUGA ...(((.(((((((.(.(((((.((....)))))))).)))))))..)))((((((((.((((........(....)........)))).))))).)))...--........... ( -34.99) >consensus UGCGUGUUGAGGUCACACACA__UGUACGCAAUGUGGCGACCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC___UACCUGAUCUCUGCGGAGA__GA___AGGUGA ...(((..((((((.((((....((....)).))))..))))))...)))((((((((..........((((((.........)))))).))))).)))................ (-25.39 = -25.52 + 0.13)


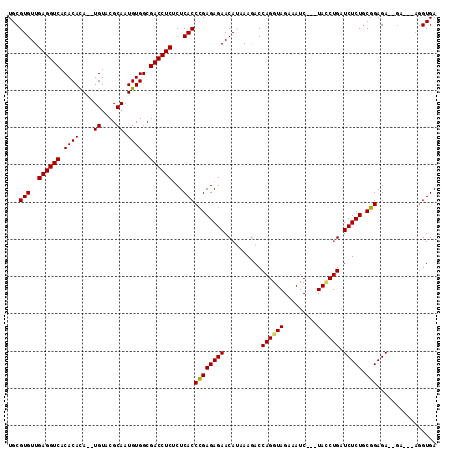
| Location | 19,041,470 – 19,041,577 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -40.21 |
| Consensus MFE | -26.99 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19041470 107 - 27905053 UCACAU---UCUCUCUCCGGAGAGAUCAGGUA---GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGGUCGCCACAUUGCGUACA--UAUGUGUGACCUCAACACGCA ......---((((.(.((((((((((((((((---(....)))))))))).......)))).)))).))))((((((((.((((((....))--.))))))))))))........ ( -45.71) >DroSec_CAF1 54085 102 - 1 UCACC--------UCUCCGCAGAGAUCAGAUA---GAUUUCUACCUGGUCUUUAUGUUCUCUCAGGUGAGAGAGGUCGCCACAUUGCGUACA--UAUGUGUGACCUCAACACGCA (((((--------(....(((((((((((.((---(....))).))))))))..)))......))))))..((((((((.((((((....))--.))))))))))))........ ( -36.60) >DroSim_CAF1 55780 105 - 1 UCACCU---UC--UCUCCGCAGAGAUCAGGUA---GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGGUCGCCACAUUGCGUACA--UGUGUGUGACCUCAACACGCA ((((((---..--....((.((((((((((((---(....))))))))))))).)).......))))))..(((((((((((((.......)--)))).))))))))........ ( -46.42) >DroEre_CAF1 57044 89 - 1 UCACCU---UC--UCUCCGCAGAGAUC-------------------GGUUUUUCUGUUCUCUCAGGUGAGUGAGGUCGCCACAAUGCGUACA--UGUGUGUGACCUCAACACGCA ((((((---..--.....(((((((..-------------------....)))))))......)))))).(((((((((((((.((....))--)))).)))))))))....... ( -34.12) >DroYak_CAF1 57417 113 - 1 UCACCUUUUUC--UCUCCGCAGAGAUCAGCUAGUUGAUGUCUACCUGGUUUUACUGUUCUCUCGGGUGAGUGAGGUCGCCACAUUGCGUACAUAUGUGUGUGACCUCAACACGCA ..........(--((((((.(((((.(((((((..(....)...)))).....))).))))))))).)))((((((((((((((((....)).))))).)))))))))....... ( -38.20) >consensus UCACCU___UC__UCUCCGCAGAGAUCAGGUA___GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGGUCGCCACAUUGCGUACA__UGUGUGUGACCUCAACACGCA ((((((...........((.(((((((((...............))))))))).)).......))))))..(((((((((((..((....))...))).))))))))........ (-26.99 = -27.73 + 0.74)



| Location | 19,041,508 – 19,041,610 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -15.53 |
| Energy contribution | -16.54 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19041508 102 + 27905053 CCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC---UACCUGAUCUCUCCGGAGAGAGA---AUGUGACACAUUAACUGAUACACC---UGUCCAAGAUGGAGG---- ..((((((.((.((((((.(.....)..(((((((....)---)))))).)))))).)).))))))---.((((.((.......)).))))..---..(((.....)))..---- ( -33.50) >DroSec_CAF1 54123 97 + 1 CCUCUCUCACCUGAGAGAACAUAAAGACCAGGUAGAAAUC---UAUCUGAUCUCUGCGGAGA--------GGUGACACAUUAACUGGUACACC---UGUCCAAGAUGGAGG---- ((((((((....)))))).......((.(((((((....)---)))))).))(((..(((.(--------((((((.((.....)))).))))---).))).))).))...---- ( -34.50) >DroSim_CAF1 55818 100 + 1 CCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC---UACCUGAUCUCUGCGGAGA--GA---AGGUGACACAUUAACUGAUACACC---UGUCCAAGAUGGAGG---- (((((.((..((((((((.(.....)..(((((((....)---)))))).))))).))).((--..---(((((...((.....))...))))---).))...)).)))))---- ( -32.70) >DroEre_CAF1 57082 88 + 1 CCUCACUCACCUGAGAGAACAGAAAAACC-------------------GAUCUCUGCGGAGA--GA---AGGUGACACAUCCACUGAGACACUUGCUGUCCAAGAUGGACGG--- .(((.(((.((..(((((...(......)-------------------..)))))..)).))--).---.((((.......))))))).......((((((.....))))))--- ( -23.50) >DroYak_CAF1 57457 112 + 1 CCUCACUCACCCGAGAGAACAGUAAAACCAGGUAGACAUCAACUAGCUGAUCUCUGCGGAGA--GAAAAAGGUGACACAUCAAGUGAUACACCUG-UGUCCAAGUUGGAGGGUUC ((((.(((..((((((((.((((........(....)........)))).))))).))).))--)....(((((.(((.....)))...))))).-...........)))).... ( -30.39) >consensus CCUCUCUCACCCGAGAGAACAUAAAGACCAGGUAGAAAUC___UACCUGAUCUCUGCGGAGA__GA___AGGUGACACAUUAACUGAUACACC___UGUCCAAGAUGGAGG____ (((((.((..((((((((..........((((((.........)))))).))))).)))...........((((...((.....))...))))..........)).))))).... (-15.53 = -16.54 + 1.01)



| Location | 19,041,508 – 19,041,610 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.86 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19041508 102 - 27905053 ----CCUCCAUCUUGGACA---GGUGUAUCAGUUAAUGUGUCACAU---UCUCUCUCCGGAGAGAUCAGGUA---GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGG ----.........((((((---.((..........)).)))).))(---(((((((((((((((((((((((---(....)))))))))).......)))).)))).))))))). ( -39.61) >DroSec_CAF1 54123 97 - 1 ----CCUCCAUCUUGGACA---GGUGUACCAGUUAAUGUGUCACC--------UCUCCGCAGAGAUCAGAUA---GAUUUCUACCUGGUCUUUAUGUUCUCUCAGGUGAGAGAGG ----..........(((.(---((((.((((.....)).))))))--------).)))..(((((((((.((---(....))).)))))))))...(((((((....))))))). ( -34.80) >DroSim_CAF1 55818 100 - 1 ----CCUCCAUCUUGGACA---GGUGUAUCAGUUAAUGUGUCACCU---UC--UCUCCGCAGAGAUCAGGUA---GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGG ----.((((((((.(((((---((((...((.....))...)))).---..--.......((((((((((((---(....))))))))))))).)))))....))))).)))... ( -37.70) >DroEre_CAF1 57082 88 - 1 ---CCGUCCAUCUUGGACAGCAAGUGUCUCAGUGGAUGUGUCACCU---UC--UCUCCGCAGAGAUC-------------------GGUUUUUCUGUUCUCUCAGGUGAGUGAGG ---.((((((.((.(((((.....))))).)))))))).....(((---((--((.((..(((((.(-------------------((.....))).)))))..)).))).)))) ( -32.10) >DroYak_CAF1 57457 112 - 1 GAACCCUCCAACUUGGACA-CAGGUGUAUCACUUGAUGUGUCACCUUUUUC--UCUCCGCAGAGAUCAGCUAGUUGAUGUCUACCUGGUUUUACUGUUCUCUCGGGUGAGUGAGG ...............((((-((((((...))))...)))))).((((...(--((((((.(((((.(((((((..(....)...)))).....))).))))))))).))).)))) ( -31.30) >consensus ____CCUCCAUCUUGGACA___GGUGUAUCAGUUAAUGUGUCACCU___UC__UCUCCGCAGAGAUCAGGUA___GAUUUCUACCUGGUCUUUAUGUUCUCUCGGGUGAGAGAGG ....(((((((((.(((((...((((...((.....))...))))...............(((((((((...............))))))))).)))))....)))))...)))) (-18.24 = -18.86 + 0.62)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:45 2006