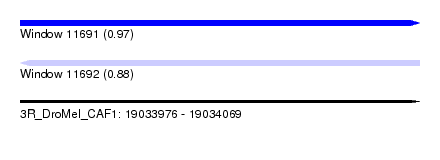
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,033,976 – 19,034,069 |
| Length | 93 |
| Max. P | 0.971285 |

| Location | 19,033,976 – 19,034,069 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19033976 93 + 27905053 AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGCGA-GGCGACG-CG-----AC--GCGCGGUGGCUGCUACUUUUACUGCCACUGCCACCGCCU------ ....((((((((.((((((((....))))))))......))))-))))..(-((-----..--(.((((((((.............)))))))))..)))..------ ( -32.42) >DroVir_CAF1 69481 79 + 1 AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGCGA------CG--------------GCG---GCAGCUACUUUUACUGCCACUGGCUGUGCCA------ ..(((((......((((((((....))))))))..)))))...------.(--------------(((---.((((((.............)))))))))).------ ( -22.72) >DroPse_CAF1 44103 92 + 1 AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGC------GACG-CG-----AC--ACGCGGUGGCAGUGGCAGUGGCGGCUGCUGCUGCUGCUU--ACAG ..(((((......((((((((....))))))))..))))).------..((-((-----..--.))))(..((((..(((((....)))))..))))..)..--.... ( -36.50) >DroGri_CAF1 57697 80 + 1 AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGCGA------CG--------------G--GCGGCAGCUACUUUUACUGCCACUGGCUGUGCCA------ ..(((((......((((((((....))))))))..)))))...------..--------------(--(((.((((((.............)))))))))).------ ( -22.42) >DroWil_CAF1 65428 102 + 1 AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGCGACGGCGGCGGCGGCAAAACAGCGGCAGCGGCUGCUACUUUUACUGCCACUGGCAGUGCCA------ ..(((((......((((((((....))))))))..)))))....(((((((((.((............)).))))))......((((((...))))))))).------ ( -35.90) >DroMoj_CAF1 50609 85 + 1 AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGCGA------CG--------------GCGGUGGCAGCUACUUUUACUGCCACUGACUGUGCCCCAG--- ..(((((......((((((((....))))))))..)))))..(------((--------------((((((((((.........))))))))).)))).......--- ( -29.10) >consensus AUCAUGUUUCGUUCAUUUUUCAAAAGAAAAAUGUUGCAUGCGA______CG______________GCGGUGGCAGCUACUUUUACUGCCACUGGCUGUGCCA______ ..(((((......((((((((....))))))))..)))))..........................(((.(((.............))).)))............... (-13.37 = -13.82 + 0.45)


| Location | 19,033,976 – 19,034,069 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -11.79 |
| Energy contribution | -12.68 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19033976 93 - 27905053 ------AGGCGGUGGCAGUGGCAGUAAAAGUAGCAGCCACCGCGC--GU-----CG-CGUCGCC-UCGCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU ------..((((((((.((.((.......)).)).))))))))((--((-----.(-((.....-.)))))))..((((((((....))))))))............. ( -35.20) >DroVir_CAF1 69481 79 - 1 ------UGGCACAGCCAGUGGCAGUAAAAGUAGCUGC---CGC--------------CG------UCGCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU ------((((...))))((((((((.......)))))---)))--------------..------...((((...((((((((....)))))))).......)))).. ( -24.00) >DroPse_CAF1 44103 92 - 1 CUGU--AAGCAGCAGCAGCAGCCGCCACUGCCACUGCCACCGCGU--GU-----CG-CGUC------GCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU .(((--(.((.((((..((((......))))..))))...((((.--..-----))-))..------)).)))).((((((((....))))))))............. ( -27.20) >DroGri_CAF1 57697 80 - 1 ------UGGCACAGCCAGUGGCAGUAAAAGUAGCUGCCGC--C--------------CG------UCGCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU ------((((...))))((((((((.......))))))))--.--------------..------...((((...((((((((....)))))))).......)))).. ( -24.00) >DroWil_CAF1 65428 102 - 1 ------UGGCACUGCCAGUGGCAGUAAAAGUAGCAGCCGCUGCCGCUGUUUUGCCGCCGCCGCCGUCGCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU ------..((((.((..(((((.((((((.((((.((....)).)))))))))).))))).)).)).))......((((((((....))))))))............. ( -32.30) >DroMoj_CAF1 50609 85 - 1 ---CUGGGGCACAGUCAGUGGCAGUAAAAGUAGCUGCCACCGC--------------CG------UCGCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU ---.((.(((...((..((((((((.......)))))))).))--------------.)------)).)).....((((((((....))))))))............. ( -24.30) >consensus ______UGGCACAGCCAGUGGCAGUAAAAGUAGCUGCCACCGC______________CG______UCGCAUGCAACAUUUUUCUUUUGAAAAAUGAACGAAACAUGAU .................(((((.((.......)).)))))............................((((...((((((((....)))))))).......)))).. (-11.79 = -12.68 + 0.89)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:37 2006