
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,026,099 – 19,026,295 |
| Length | 196 |
| Max. P | 0.789551 |

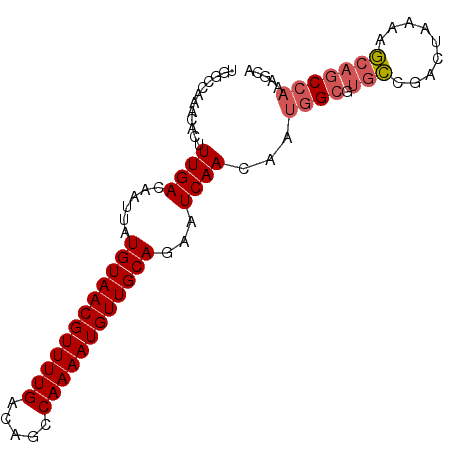
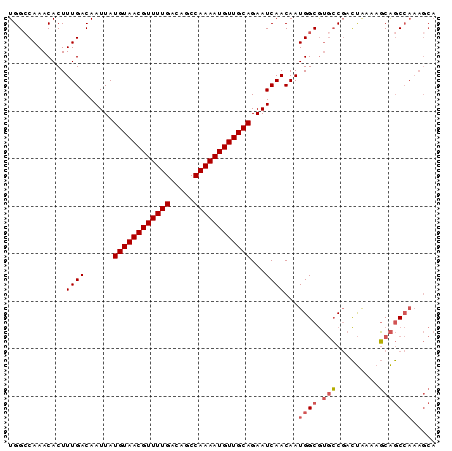
| Location | 19,026,099 – 19,026,189 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19026099 90 + 27905053 UGGCCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCGUGCCGACUAAAAGCAGCCAAAGCA ..((........((((......((((((((((((.....))))))))))))...))))...((((.(((.........)))))))..)). ( -22.90) >DroSec_CAF1 38908 90 + 1 UGGCCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCGUGCCGACUAAAAGCAGCCAAAGCA ..((........((((......((((((((((((.....))))))))))))...))))...((((.(((.........)))))))..)). ( -22.90) >DroSim_CAF1 40566 90 + 1 UGGCCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCGUGCCGACUAAAAGCAGCCAAAGCA ..((........((((......((((((((((((.....))))))))))))...))))...((((.(((.........)))))))..)). ( -22.90) >DroEre_CAF1 41391 90 + 1 UGGGCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCGUGCCGACUAAAAGCAGCCAAAGCA ...((.......((((......((((((((((((.....))))))))))))...))))...((((.(((.........)))))))..)). ( -23.80) >DroYak_CAF1 41681 90 + 1 UGGCCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCGUGCCGACUAAAAGCAGCCAAAGCA ..((........((((......((((((((((((.....))))))))))))...))))...((((.(((.........)))))))..)). ( -22.90) >DroAna_CAF1 39522 85 + 1 UGGCCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCAACACCAUCGACAUCGACAU----- ..((((......((((......((((((((((((.....))))))))))))...))))...)))).......(((....)))...----- ( -20.20) >consensus UGGCCAAACACUUUGACAAUUAUGUAACGUUUUGACAGCCAAAAUGUUGCAGAAUCAACAAUGGCGUGCCGACUAAAAGCAGCCAAAGCA ............((((......((((((((((((.....))))))))))))...))))...((((.(((.........)))))))..... (-17.78 = -18.33 + 0.56)

| Location | 19,026,099 – 19,026,189 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19026099 90 - 27905053 UGCUUUGGCUGCUUUUAGUCGGCACGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGGCCA .....(((((((....((((((((......))))))))..))(((((((((((....................))))))))))).))))) ( -23.95) >DroSec_CAF1 38908 90 - 1 UGCUUUGGCUGCUUUUAGUCGGCACGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGGCCA .....(((((((....((((((((......))))))))..))(((((((((((....................))))))))))).))))) ( -23.95) >DroSim_CAF1 40566 90 - 1 UGCUUUGGCUGCUUUUAGUCGGCACGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGGCCA .....(((((((....((((((((......))))))))..))(((((((((((....................))))))))))).))))) ( -23.95) >DroEre_CAF1 41391 90 - 1 UGCUUUGGCUGCUUUUAGUCGGCACGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGCCCA .((((((((.((.....)).((((.((((.(((((......)))))...))))))))................))))))))......... ( -23.60) >DroYak_CAF1 41681 90 - 1 UGCUUUGGCUGCUUUUAGUCGGCACGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGGCCA .....(((((((....((((((((......))))))))..))(((((((((((....................))))))))))).))))) ( -23.95) >DroAna_CAF1 39522 85 - 1 -----AUGUCGAUGUCGAUGGUGUUGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGGCCA -----..(((((.(((((..(((....)))..)))))))...(((((((((((....................))))))))))).))).. ( -20.25) >consensus UGCUUUGGCUGCUUUUAGUCGGCACGCCAUUGUUGAUUCUGCAACAUUUUGGCUGUCAAAACGUUACAUAAUUGUCAAAGUGUUUGGCCA ((((..(((((....))))))))).((((.(((((......)))))...)))).((((((((...(((....)))....)).)))))).. (-20.40 = -20.27 + -0.13)

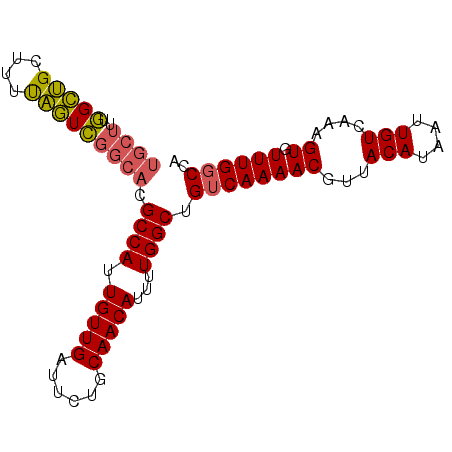

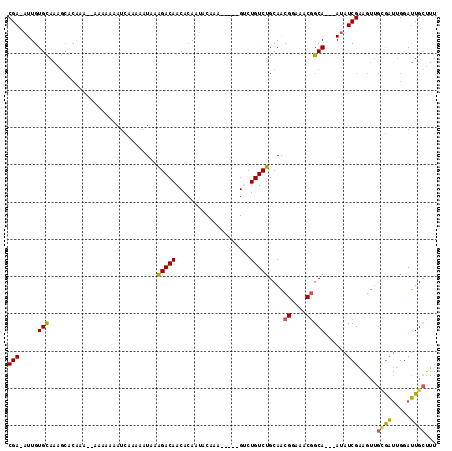
| Location | 19,026,189 – 19,026,295 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -11.84 |
| Energy contribution | -11.53 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19026189 106 - 27905053 CGA-AUUGUGCAAAGCCCAAAAAAAAAAAAUCAAAAAUAAAGACAACACAAUACAAA-----GUCUGUCUGCAACGGAAACGGCA---AUAUCGAAGUUGCGAUUGGAUUGCUUU ...-.....((((...((((....................(((((..((........-----)).)))))....((....))(((---((......)))))..)))).))))... ( -18.40) >DroSec_CAF1 38998 101 - 1 CGA-AUUGUGCAAAGCACAAA-----AAAAUCAAAAAUAAAGACAACACAAUACAAA-----GACUGUCUGCAACGGAAACGGCA---AUAUCGAAGUUGCGAUUGGAUUGCUUU ...-.((((((...)))))).-----............((((.(((..((((.(...-----............((....))(((---((......))))))))))..))))))) ( -20.60) >DroSim_CAF1 40656 101 - 1 CGA-AUUGUGCAAAGCACAAA-----AAAAUCAAAAAUAAAGACAACACAAUACAAA-----GGCUGUCUGCAACGGAAACGGCA---AUAUCGAAGUUGCGAUUGGAUUGCUUU ...-.((((((...)))))).-----............((((.(((..((((.(...-----.((.....))..((....))(((---((......))))))))))..))))))) ( -21.70) >DroEre_CAF1 41481 106 - 1 CGA-AUUGUGCAAAGCACAAACAAAAAAAAUCAAAAAUAAAGACAACACAAUACAAC-----GUCUGUCUGCAACGGAAACGGCA---AUAUCGAAGUUGCGAUUGGAUUGCUUU ...-.((((((...))))))..................((((.(((..((((.((((-----.((.((.(((..((....)))))---..)).)).))))..))))..))))))) ( -21.60) >DroYak_CAF1 41771 103 - 1 CGA-AUUGUGCAAAGCUCAA---AAAAAAAUCAAAAAUAAAGACAACACAAUACAAA-----GUCUGUCUGCAACGGAAACGGCA---AUAUCGAAGUUGUGAUUGGAUUGCUUU ...-.(((.((...)).)))---...............((((.(((..((((((((.-----.((.((.(((..((....)))))---..)).))..)))).))))..))))))) ( -18.30) >DroPer_CAF1 35259 100 - 1 CGAAAUUGUGUAA----------AGAAAAAUAAAAGUU--GGACAACAAAAAGCUAACGUCUGUCUGUCUGCAGCGGAAACAGCAGACAGAUCGGUG---GAGGAGGACUGUGUG ......(((.(((----------.............))--).)))(((.....((..(..(.((((((((((...(....).)))))))))).)..)---..)).....)))... ( -23.92) >consensus CGA_AUUGUGCAAAGCACAAA__AAAAAAAUCAAAAAUAAAGACAACACAAUACAAA_____GUCUGUCUGCAACGGAAACGGCA___AUAUCGAAGUUGCGAUUGGAUUGCUUU (((.....(((.............................(((((....................)))))....((....)))))......))).....((((.....))))... (-11.84 = -11.53 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:34 2006