
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,023,989 – 19,024,087 |
| Length | 98 |
| Max. P | 0.813792 |

| Location | 19,023,989 – 19,024,087 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.78 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -6.78 |
| Energy contribution | -6.53 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19023989 98 + 27905053 --UUGC-----CUCGAAAUUCCCCAGAGAGUGAUCAAUUUUCUGCU--ACAAAAACAGUUGGAAGGUUCAUAUCUGUGGCGGGGGGGGGAGCAGGAAAG--UAGGUAAA --((((-----(((....((((((...((....))...(..(((((--((.....(....)..((((....)))))))))))..))))))).......)--.)))))). ( -28.30) >DroSec_CAF1 36882 88 + 1 --UUGA-----AUCGAAACUCCCCAGAGAGUGAUCAAUUUUCUGCU--AC------AGUUGGAAGGUUGAAAUCUGUGG----GGGGCACACAGGAAAG--UAGGUAAA --....-----.......(((((((.(((....((((((((((((.--..------.).)))))))))))..))).)))----))))............--........ ( -25.90) >DroSim_CAF1 38531 92 + 1 --UUGA-----AUCGAAACUCCCCAGAGAGUGAUCAAUUUUCUGCU--ACAAAAACAGUUGGAAGGUUCAAAUCUGUGG----GGGGCA--CAGCAAAG--UAGGUAAA --....-----.......(((((((.(((.(((....(((((.(((--........))).)))))..)))..))).)))----))))..--........--........ ( -22.60) >DroEre_CAF1 39257 90 + 1 --UUGC-----CUGCAAACUCCCCAGAGAGUGAUCAAUUUUCUGCU--ACAAAAACAGUUUGAAGGUUCAAAUCUGUGG----G----GGGCAGGAGAG--UAGGUAGC --((((-----((((..((((......))))......((((((((.--.(....(((((((((....))))).))))..----)----..)))))))))--))))))). ( -31.90) >DroYak_CAF1 39650 79 + 1 --UUGC-----CUCGCAACUCCCCAGAGAGUGAUCAAUUUUCUGCU--ACAAAAACAGU---AAGGUAAA--------G----U----AAACAGGAAAG--UAGGUAAA --((((-----((.((...(((.((((((((.....)))))))).(--((.......))---).......--------.----.----.....)))..)--))))))). ( -15.40) >DroAna_CAF1 37607 105 + 1 CCUUGAAACUUCUCCAUAUUCCCCGGAGUGUGAUCAAUAUUCUUCGGAAAAAAAAUAGUUGCAAGACCAUCAUAUGCCC----CGGGGACCAAGGAAAGGCAAGGUAAA (((((...(((.(((....(((((((.(((((((.....(((....)))........(((....))).)))))))...)----))))))....)))))).))))).... ( -34.50) >consensus __UUGA_____CUCGAAACUCCCCAGAGAGUGAUCAAUUUUCUGCU__ACAAAAACAGUUGGAAGGUUCAAAUCUGUGG____GGGG_AAACAGGAAAG__UAGGUAAA .......................((((((((.....))))))))................................................................. ( -6.78 = -6.53 + -0.25)

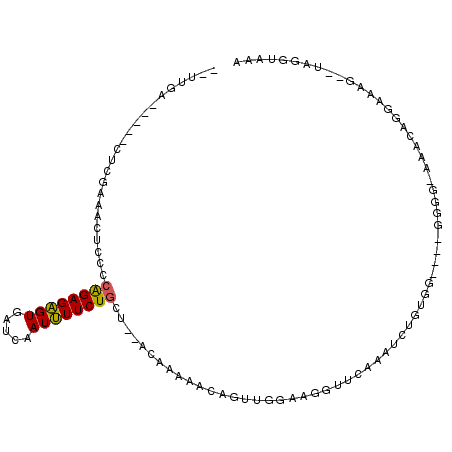
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:32 2006