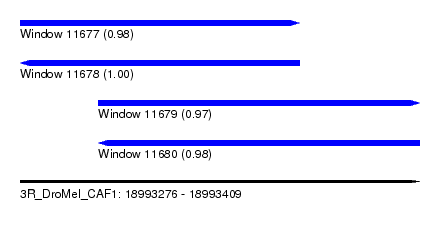
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,993,276 – 18,993,409 |
| Length | 133 |
| Max. P | 0.997377 |

| Location | 18,993,276 – 18,993,369 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -28.10 |
| Energy contribution | -27.54 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18993276 93 + 27905053 GG--------GGUGUUACUAUGGUCGAAACCAUUGUUGUCAAUUUAAGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCUCACGAAA .(--------((((..((.(((((....))))).))...)...........((((..(((((((((.((....)).)))))))))---))))..))))...... ( -29.30) >DroSec_CAF1 6949 94 + 1 GGG-------GGUGUUACUAUGGUCGAAACCAUUGUUGUCAAUUCACGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCUCACGAAA ...-------.(((...(((((((....)))))((((((......))))))((((..(((((((((.((....)).)))))))))---))))))...))).... ( -29.80) >DroSim_CAF1 6941 93 + 1 AC--------UAUGGUACUAUGGUCGAAACCAUUGUUGUCAAUUUACGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCUCACGAAA ((--------((((....)))))).((..((..((((((......))))))((((..(((((((((.((....)).)))))))))---))))))..))...... ( -30.40) >DroEre_CAF1 8089 98 + 1 GGGA------GGAGUUACUAUGGUCGAAACCAUUGUUGUCAGUUUACGAUACUGCCACCGCUGCCAUUUGAUUGAUUGGCAGCGGGCGGCAGUGGCCCAUGGAU (((.------.((...((.(((((....))))).))..)).........(((((((.(((((((((.((....)).)))))))))..))))))).)))...... ( -41.90) >DroYak_CAF1 7293 101 + 1 GGGGGUGGGGGAUGUUACUAUGGUCGCAACCAUUGUUGUCAAUUUACGAUACUGCUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCGCGCGAAA ....(((.(((.(((.((....)).))).))..((((((......))))))((((..(((((((((.((....)).)))))))))---))))...).))).... ( -33.50) >consensus GGG_______GGUGUUACUAUGGUCGAAACCAUUGUUGUCAAUUUACGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG___GCAGGGGCUCACGAAA ................((.(((((....))))).))((((.......))))((((..(((((((((.((....)).)))))))))...))))............ (-28.10 = -27.54 + -0.56)



| Location | 18,993,276 – 18,993,369 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18993276 93 - 27905053 UUUCGUGAGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCUUAAAUUGACAACAAUGGUUUCGACCAUAGUAACACC--------CC ....(((.....(((.---(((((((((..........)))))))))...)))...............((.(((((....))))).))..))).--------.. ( -24.40) >DroSec_CAF1 6949 94 - 1 UUUCGUGAGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCGUGAAUUGACAACAAUGGUUUCGACCAUAGUAACACC-------CCC .(((((((....(((.---(((((((((..........)))))))))...)))..)))))))......((.(((((....))))).))......-------... ( -26.00) >DroSim_CAF1 6941 93 - 1 UUUCGUGAGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCGUAAAUUGACAACAAUGGUUUCGACCAUAGUACCAUA--------GU .((..(((....(((.---(((((((((..........)))))))))...)))..)))..))......((.(((((....))))).))......--------.. ( -23.60) >DroEre_CAF1 8089 98 - 1 AUCCAUGGGCCACUGCCGCCCGCUGCCAAUCAAUCAAAUGGCAGCGGUGGCAGUAUCGUAAACUGACAACAAUGGUUUCGACCAUAGUAACUCC------UCCC ....((((...(((((((((.(((((((..........)))))))))))))))).(((.(((((.........)))))))))))).........------.... ( -37.90) >DroYak_CAF1 7293 101 - 1 UUUCGCGCGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAGCAGUAUCGUAAAUUGACAACAAUGGUUGCGACCAUAGUAACAUCCCCCACCCCC ..(((((.(((.((((---(((((((((..........)))))))))..))))........((((....))))))))))))....................... ( -28.70) >consensus UUUCGUGAGCCCCUGC___CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCGUAAAUUGACAACAAUGGUUUCGACCAUAGUAACACC_______CCC ....((((....(((....(((((((((..........)))))))))...)))..)))).........((.(((((....))))).))................ (-22.90 = -22.58 + -0.32)



| Location | 18,993,302 – 18,993,409 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18993302 107 + 27905053 GUUGUCAAUUUAAGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCUCACGAAAAGGGAUGGAACUGAUUUGGAGCUAGUUUACUUUCAUUUUU ..((((.......))))((((..(((((((((.((....)).)))))))))---)))).(((((.((((..((.......))..)))))))))................. ( -32.20) >DroSec_CAF1 6976 107 + 1 GUUGUCAAUUCACGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCUCACGAAAAGGGAUUGAACUAAGUUGAAGCUACUUUACUUUAGUUUUU ....(((((((......((((..(((((((((.((....)).)))))))))---))))(......)......)))))))((((((.(((((...))))).)))))).... ( -31.50) >DroSim_CAF1 6967 106 + 1 GUUGUCAAUUUACGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCUCACGAAAAGGGGUUGAACUGAUUUAAAGCUACUUUACUUUA-UUUUU ...((((.(((......((((..(((((((((.((....)).)))))))))---)))).(((((.(.....))))))))).)))).(((((........)))))-..... ( -27.30) >DroEre_CAF1 8117 92 + 1 GUUGUCAGUUUACGAUACUGCCACCGCUGCCAUUUGAUUGAUUGGCAGCGGGCGGCAGUGGCCCAUGGAUAGGGAUUCAACUGGCUUAAAGC------------------ ...(((((((.....(((((((.(((((((((.((....)).)))))))))..))))))).(((.......)))....))))))).......------------------ ( -42.70) >DroYak_CAF1 7327 104 + 1 GUUGUCAAUUUACGAUACUGCUACUGCUGCCAUUUGAUUGAUUGGCAGCAG---GCAGGGGCGCGCGAAAGGGGAUUCAACAGGCUUGCAGCUCC--UAGUUUA-CUUCU (((((......))))).((((..(((((((((.((....)).)))))))))---))))(((.((((((....(.......)....)))).)).))--)......-..... ( -32.20) >consensus GUUGUCAAUUUACGAUACUGUUACUGCUGCCAUUUGAUUGAUUGGCAGCAG___GCAGGGGCUCACGAAAAGGGAUUGAACUGACUUGAAGCUACUUUACUUUA_UUUUU ..((((.......))))((((..(((((((((.((....)).)))))))))...)))).(((...(((..((........))...)))..)))................. (-24.66 = -24.30 + -0.36)

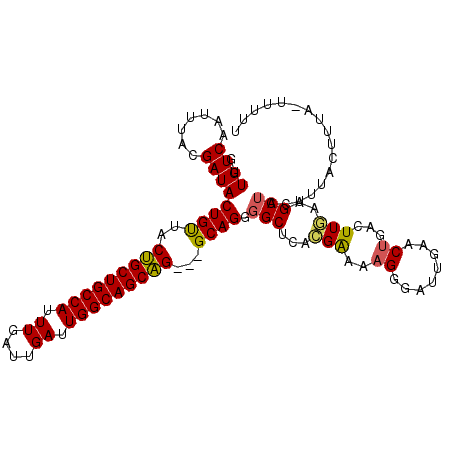

| Location | 18,993,302 – 18,993,409 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18993302 107 - 27905053 AAAAAUGAAAGUAAACUAGCUCCAAAUCAGUUCCAUCCCUUUUCGUGAGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCUUAAAUUGACAAC ....(((((((......((((.......)))).......)))))))(((...(((.---(((((((((..........)))))))))...)))...)))........... ( -20.52) >DroSec_CAF1 6976 107 - 1 AAAAACUAAAGUAAAGUAGCUUCAACUUAGUUCAAUCCCUUUUCGUGAGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCGUGAAUUGACAAC ...(((((((((......))).....))))))......(..(((((((....(((.---(((((((((..........)))))))))...)))..)))))))..)..... ( -23.80) >DroSim_CAF1 6967 106 - 1 AAAAA-UAAAGUAAAGUAGCUUUAAAUCAGUUCAACCCCUUUUCGUGAGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCGUAAAUUGACAAC .....-((((((......))))))..((((((...........(....)...(((.---(((((((((..........)))))))))...))).......)))))).... ( -23.30) >DroEre_CAF1 8117 92 - 1 ------------------GCUUUAAGCCAGUUGAAUCCCUAUCCAUGGGCCACUGCCGCCCGCUGCCAAUCAAUCAAAUGGCAGCGGUGGCAGUAUCGUAAACUGACAAC ------------------((.....))(((((....(((.......)))..(((((((((.(((((((..........))))))))))))))))......)))))..... ( -37.70) >DroYak_CAF1 7327 104 - 1 AGAAG-UAAACUA--GGAGCUGCAAGCCUGUUGAAUCCCCUUUCGCGCGCCCCUGC---CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAGCAGUAUCGUAAAUUGACAAC ....(-(....((--.(((((((..((.(((.(((......)))))).))......---(((((((((..........)))))))))..))))).)).)).....))... ( -28.90) >consensus AAAAA_UAAAGUAAAGUAGCUUCAAAUCAGUUCAAUCCCUUUUCGUGAGCCCCUGC___CUGCUGCCAAUCAAUCAAAUGGCAGCAGUAACAGUAUCGUAAAUUGACAAC ...........................(((((...........(....)...(((....(((((((((..........)))))))))...))).......)))))..... (-17.72 = -17.28 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:26 2006