
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,988,169 – 18,988,278 |
| Length | 109 |
| Max. P | 0.643740 |

| Location | 18,988,169 – 18,988,278 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -18.72 |
| Energy contribution | -21.38 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18988169 109 - 27905053 UAUUUGCAUUAUUUUCACGCCAUU---------CCCGCUGUUCACACAGCUGUUGGCCAUUUGGAUGGGGGCCAGUUCUCUGGCAGACCAUAAAGAUUGGCUAGAACAAGAACCAAAG ..((((.......(((........---------...(((((....)))))...((((((.((..((((..(((((....)))))...))))...)).))))))))).......)))). ( -31.54) >DroVir_CAF1 9367 100 - 1 -ACCGGCCACAGCUGUUUGCCAUUCUCAUCUG--GCGCGGUGC--ACGGAUUUUUGUCUUUUGGAUGUAAGCCAAUU-UCAAAUAGACCAUAAAAUAC-------ACAAG-A----AG -.(((((....(((((..((((........))--)))))))))--.)))(((((.((((((((((...........)-))))).))))...)))))..-------.....-.----.. ( -21.50) >DroSec_CAF1 1825 108 - 1 UAUUUGCAUUAUUUUCACGCCAUU---------CCCGCUGUUCUCGCAGCUGUUGGCCAUUUGGAUGGUGGCCAGUUCUCUGGCAGACCAUAUAGAAUGGCUAGAACAAG-ACCAAAG ..((((..................---------...(((((....)))))..((((((((((..(((((.(((((....)))))..)))))...))))))))))......-..)))). ( -35.40) >DroSim_CAF1 1807 108 - 1 UAUUUGCAUUAUUUUCACGCCAUU---------CCCGCUGUUCUCGCAGCUGUUGGCCAUUUGGAUGGUGGCCAGUUCUCUGGCAGACCAUCUAGAAUGGCUAGAACAAG-ACCAAAG ..((((..................---------...(((((....)))))..(((((((((((((((((.(((((....)))))..))))))))).))))))))......-..)))). ( -38.30) >DroEre_CAF1 1828 107 - 1 UAUUUGCAUUAUUUUCACGGCUUU---------CCCGCUGUUCUCGCAGCUGUUGGCCAUUUGGACGGGGGCCAGUUCUCUGGCAGACCAGAAAGUAUGGCUAGAACAAG-AC-CAAG .........(((((((..((((..---------(..(((((....))))).)..))))........((..(((((....)))))...)).)))))))(((((......))-.)-)).. ( -32.80) >DroYak_CAF1 1799 117 - 1 UAUGUUCAUUAUUUUCACGCCUUUCCCGUGUUUCCCGCUGCUCGCGCAGCUGUUGGCCAUUUGGAUGGGGGCCAGUUCUCUGGCAGACCAGAAACUUUGGCUAGAACAAG-ACCCAAG ..(((((........((((.......))))......(((((....)))))...((((((......(((..(((((....)))))...))).......)))))))))))..-....... ( -37.22) >consensus UAUUUGCAUUAUUUUCACGCCAUU_________CCCGCUGUUCUCGCAGCUGUUGGCCAUUUGGAUGGGGGCCAGUUCUCUGGCAGACCAUAAAGAAUGGCUAGAACAAG_ACCAAAG ....................................(((((....)))))..((((((((....((((..(((((....)))))...)))).....)))))))).............. (-18.72 = -21.38 + 2.67)

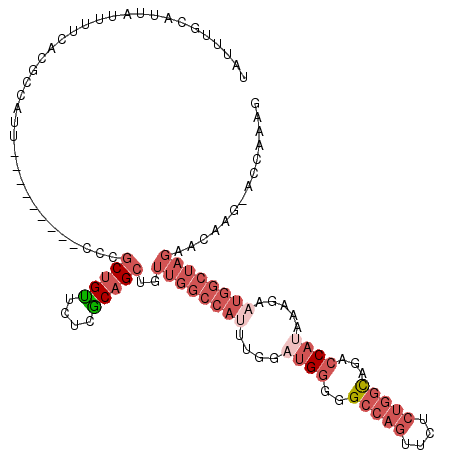
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:22 2006