
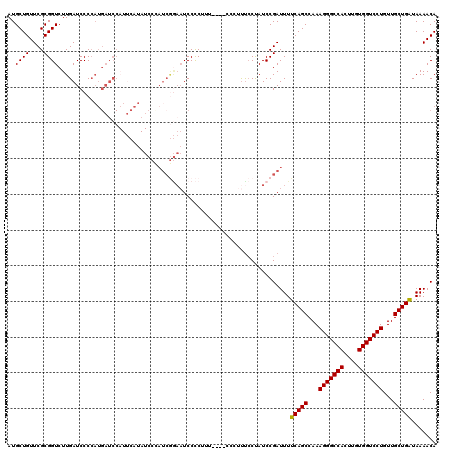
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,979,541 – 18,979,664 |
| Length | 123 |
| Max. P | 0.928180 |

| Location | 18,979,541 – 18,979,657 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.50 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
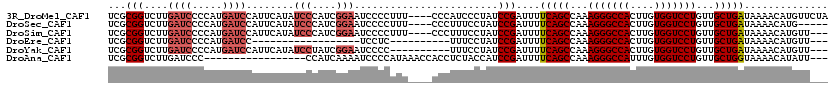
>3R_DroMel_CAF1 18979541 116 + 27905053 AUGCUGUUCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU----CCCAUCCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACA ..((((....))))...((((.....))))............((((((.........----..........))))))..(((((...(((((((....)))))))...)))))....... ( -28.31) >DroSec_CAF1 90523 116 + 1 AUGCUGUUCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU----CCCUUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACA ..((((....))))...((((.....))))............((((((.........----..........))))))..(((((...(((((((....)))))))...)))))....... ( -28.31) >DroSim_CAF1 92335 116 + 1 AUGCUGUUCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU----CCCUUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACA ..((((....))))...((((.....))))............((((((.........----..........))))))..(((((...(((((((....)))))))...)))))....... ( -28.31) >DroEre_CAF1 91116 92 + 1 AUGCUGUUCGCGGUCUUGAUCCCCAUGAUCC------------------UCCUC----------UUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACA ..((.....))((....((((.....)))).------------------.))..----------...............(((((...(((((((....)))))))...)))))....... ( -23.90) >DroYak_CAF1 93721 110 + 1 AUGCUGUUCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCUAUCGGAAUCCCC----------UUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACA ..((((....))))...((((.....))))............((((((......----------.......))))))..(((((...(((((((....)))))))...)))))....... ( -28.62) >DroAna_CAF1 93440 103 + 1 AUGCUGUUCGCGGUCUUGAUCCC-----------------CCAUCAAAAUCCCCAUAAACCACCUCUACCAUCCGAUUUUCAGCCAAAGGGCCAUUUGUGGUCCUGUUGCUGGUAAAACA ..((.....))(((.(((((...-----------------..))))).)))............................(((((...(((((((....)))))))...)))))....... ( -21.80) >consensus AUGCUGUUCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU____CCCUUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACA ..((((....)))).................................................................(((((...(((((((....)))))))...)))))....... (-21.64 = -21.50 + -0.14)


| Location | 18,979,548 – 18,979,664 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.88 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18979548 116 + 27905053 UCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU----CCCAUCCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACAUGUUCUA ...(((....(((....((((.....))))........((((......))----)).))).....)))....(((((...(((((((....)))))))...))))).............. ( -28.00) >DroSec_CAF1 90530 111 + 1 UCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU----CCCUUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACAUG----- ...(((....((.....((((.....))))........((((......))----))...))....)))....(((((...(((((((....)))))))...))))).........----- ( -27.90) >DroSim_CAF1 92342 113 + 1 UCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU----CCCUUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACAUGUU--- ...(((....((.....((((.....))))........((((......))----))...))....)))....(((((...(((((((....)))))))...)))))...........--- ( -27.90) >DroEre_CAF1 91123 89 + 1 UCGCGGUCUUGAUCCCCAUGAUCC------------------UCCUC----------UUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACAUGUU--- ....((....((((.....)))).------------------.))..----------...............(((((...(((((((....)))))))...)))))...........--- ( -23.00) >DroYak_CAF1 93728 107 + 1 UCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCUAUCGGAAUCCCC----------UUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACAUGUU--- ...(((....(((....((((.....)))).....)))((((.....----------.))))...)))....(((((...(((((((....)))))))...)))))...........--- ( -28.30) >DroAna_CAF1 93447 100 + 1 UCGCGGUCUUGAUCCC-----------------CCAUCAAAAUCCCCAUAAACCACCUCUACCAUCCGAUUUUCAGCCAAAGGGCCAUUUGUGGUCCUGUUGCUGGUAAAACAUAUU--- ..(.((..(((((...-----------------..)))))...)).).........................(((((...(((((((....)))))))...)))))...........--- ( -21.10) >consensus UCGCGGUCUUGAUCCCCAUGAUCCAUUCAUAUCCCAUCGGAAUCCCCUUU____CCCUUUCCUAUCCGAUUUUCAGCCAAAGGGCCACUUGUGGUCCUGUUGCUGAUAAAACAUGUU___ ...(((....((((.....))))........(((....)))........................)))....(((((...(((((((....)))))))...))))).............. (-20.73 = -21.88 + 1.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:21 2006