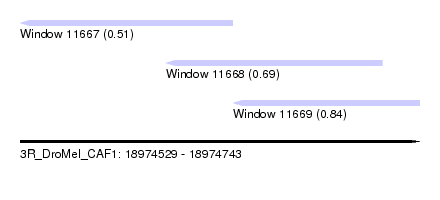
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
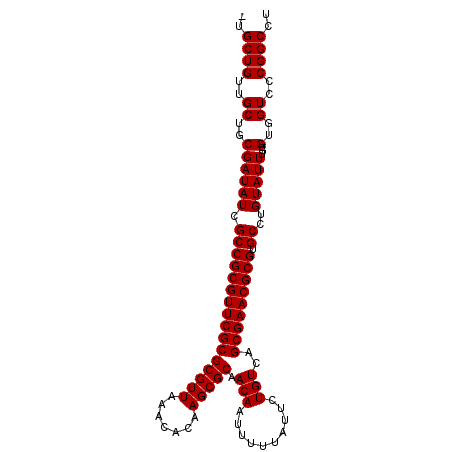
| Location | 18,974,529 – 18,974,743 |
| Length | 214 |
| Max. P | 0.837362 |

| Location | 18,974,529 – 18,974,643 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18974529 114 - 27905053 CAAACUGGACUGGACCGUGGCUCCAUGGGCUCCUGCUGCCUGCUGCCUGCUGCCUGCCAUCCUUACCAUUCCUGCCACCUCCACCUCCUAUCUCCU-UCUCUACCUUCUGAUCUU ......(((.((((..(((((...((((......((.((..((.....)).))..))........))))....))))).))))..)))........-.................. ( -23.44) >DroSec_CAF1 85579 99 - 1 CAAACUGGACUGGACCGUGGCUCCAUGGGCUCCUGCU--------------GCCUGCCAUCCUUACCAUUCCUGCCACCUCCACCUCC-AUCUCCU-GCUCUGCCUUCUGAUCUU .....((((.((((..(((((...((((......((.--------------....))........))))....))))).))))..)))-)......-.................. ( -20.34) >DroSim_CAF1 87372 106 - 1 CAAACUGGACUGGACCGUGGCUCCAUGGGCUCCUGCUGCCU-------GCUGCCUGCCAUCCUUACCAUUCCUGCCACCUCCACCUCC-AUCCUCU-GCUCUGCCUUCUGAUCUU .....((((.((((..(((((...((((((....)).....-------((.....))........))))....))))).))))..)))-)......-.................. ( -22.10) >DroEre_CAF1 86171 100 - 1 CAAACUGGACUGGACCGUGGCUCCAUGGGCUCCUGCU--------------GCCUGCCAUCCUUACCAUUCCUACCACCUCCACCUCC-AUCUCCUUCCUCUGCCUUCUGAUCUU ......(((.(((...(((((.....((((.......--------------))))))))).....))).)))................-.......................... ( -17.20) >DroYak_CAF1 88744 101 - 1 CAAACUGGACUGGACCGUGGCUCCAUGGGCUCCUGCUGCCU-------GCUGCCUGCCAUCCUUACCAUUCCAACCACC------UCC-AUCUCCUUCCUCUGCCUUCUGAUCUU .....((((.(((...(((((..(((((((.......))))-------).))...))))).....))).))))......------...-.......................... ( -20.80) >consensus CAAACUGGACUGGACCGUGGCUCCAUGGGCUCCUGCUGCCU_______GCUGCCUGCCAUCCUUACCAUUCCUGCCACCUCCACCUCC_AUCUCCU_CCUCUGCCUUCUGAUCUU ......(((.(((..((((....))))(((....((.((.........)).))..))).......))).)))........................................... (-16.28 = -16.48 + 0.20)

| Location | 18,974,607 – 18,974,723 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -37.14 |
| Energy contribution | -37.50 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18974607 116 - 27905053 CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCUCAAACUGGACUGGACCGU----GGCUCCAUGGGCUCCUGC ((((((((((((((.......))))).(((..........)))..))))))))).((((..........(((((.((((.......)).)))))))((----(....)))))))...... ( -39.80) >DroSec_CAF1 85642 116 - 1 CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCUCAAACUGGACUGGACCGU----GGCUCCAUGGGCUCCUGC ((((((((((((((.......))))).(((..........)))..))))))))).((((..........(((((.((((.......)).)))))))((----(....)))))))...... ( -39.80) >DroSim_CAF1 87442 116 - 1 CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCUCAAACUGGACUGGACCGU----GGCUCCAUGGGCUCCUGC ((((((((((((((.......))))).(((..........)))..))))))))).((((..........(((((.((((.......)).)))))))((----(....)))))))...... ( -39.80) >DroEre_CAF1 86235 116 - 1 CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCUCAAACUGGACUGGACCGU----GGCUCCAUGGGCUCCUGC ((((((((((((((.......))))).(((..........)))..))))))))).((((..........(((((.((((.......)).)))))))((----(....)))))))...... ( -39.80) >DroYak_CAF1 88809 116 - 1 CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCUCAAACUGGACUGGACCGU----GGCUCCAUGGGCUCCUGC ((((((((((((((.......))))).(((..........)))..))))))))).((((..........(((((.((((.......)).)))))))((----(....)))))))...... ( -39.80) >DroAna_CAF1 88242 120 - 1 CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGUCUUCAAACUGGACUGGACUGGGAAGGGGACCCUUGGCUCCUUU ((((((((((((((.......))))).(((..........)))..))))))))).(((........(.((((((.((((.......)))).)))))).)((((....)))))))...... ( -41.60) >consensus CGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCUCAAACUGGACUGGACCGU____GGCUCCAUGGGCUCCUGC ((((((((((((((.......))))).(((..........)))..))))))))).((((((........(((((.(........).)))))(((((......)).)))))))))...... (-37.14 = -37.50 + 0.36)


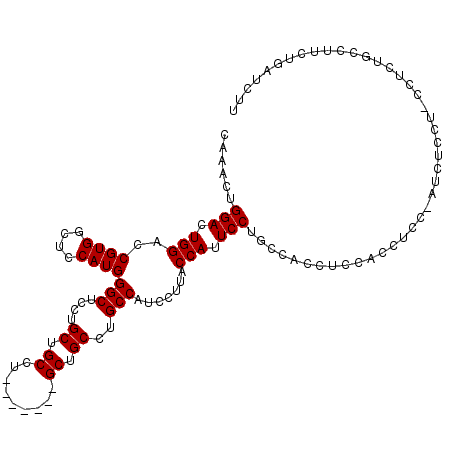
| Location | 18,974,643 – 18,974,743 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 99.50 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -35.80 |
| Energy contribution | -35.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18974643 100 - 27905053 -UGCUGUUGCUGCGAUAUCGCCGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCU -.((((..((..((((((.((((((((((((((((.......))))).(((..........)))..))))))))).))..)))))...)..))..)))).. ( -35.80) >DroSec_CAF1 85678 100 - 1 -UGCUGUUGCUGCGAUAUCGCCGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCU -.((((..((..((((((.((((((((((((((((.......))))).(((..........)))..))))))))).))..)))))...)..))..)))).. ( -35.80) >DroSim_CAF1 87478 100 - 1 -UGCUGUUGCUGCGAUAUCGCCGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCU -.((((..((..((((((.((((((((((((((((.......))))).(((..........)))..))))))))).))..)))))...)..))..)))).. ( -35.80) >DroYak_CAF1 88845 101 - 1 UUGCUGUUGCUGCGAUAUCGCCGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCU ..((((..((..((((((.((((((((((((((((.......))))).(((..........)))..))))))))).))..)))))...)..))..)))).. ( -35.80) >consensus _UGCUGUUGCUGCGAUAUCGCCGCGUUCGCGCGUUAAACACAAGCGCAACAAUUUUUAUUCUGUCAGCGAACGCGUGCCUGUAUUUGUGUGGUCCCGGCCU ..((((..((..((((((.((((((((((((((((.......))))).(((..........)))..))))))))).))..)))))...)..))..)))).. (-35.80 = -35.80 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:15 2006