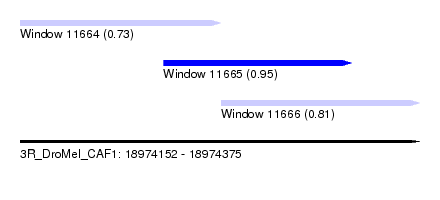
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,974,152 – 18,974,375 |
| Length | 223 |
| Max. P | 0.949769 |

| Location | 18,974,152 – 18,974,264 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -31.70 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
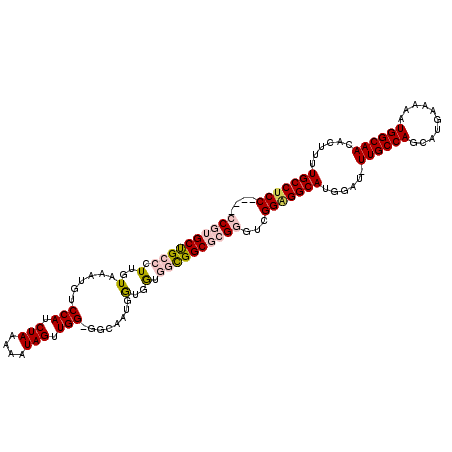
>3R_DroMel_CAF1 18974152 112 + 27905053 GCAGGAGAACAAGUUUCGAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGGACACGAAUCGAGAUCGAGGCGAAAGCCUGA-CGCUGAUGCAGAUCUC------- (((((....)..((((((.((....((((.....))))...))...)))))).))))..................((((((.((((....))))..-.((....)).))))))------- ( -36.10) >DroSec_CAF1 85207 112 + 1 GCAGGAGAACAAGUUUCGAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGGACACGAAUCGGGAUCGAGGCGAAAGCCUGA-CGCUGAUGCAGAUCUC------- ....((((....((((((.((....((((.....))))...))...)))))).((((......((....)).(((((..((.((((....))))))-..))))))))).))))------- ( -36.70) >DroSim_CAF1 87000 112 + 1 GCAGGAGAACAAGUUUCGAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGGACACGAAUCGGGAUCGAGGCGAAAGCCUGA-CGCUGAUGCAGAUCUC------- ....((((....((((((.((....((((.....))))...))...)))))).((((......((....)).(((((..((.((((....))))))-..))))))))).))))------- ( -36.70) >DroEre_CAF1 85804 112 + 1 GCAAGAGAACAAGUUUCGAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGGACACGAAUCGAGAUCGAGGCGAAAGCCUGA-CGCUGAUGCAGAUCUC------- ......((....((.((..(((....(((((((..((..(...)..))..)))).)))...))).)).))...))((((((.((((....))))..-.((....)).))))))------- ( -35.90) >DroYak_CAF1 88369 110 + 1 --CAGAGAACAAGUUUCGAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGGACACGAAUCGAGAUCGAGGCGAAAGCCUGA-CGCUGAUGCAGAUCUC------- --....((....((.((..(((....(((((((..((..(...)..))..)))).)))...))).)).))...))((((((.((((....))))..-.((....)).))))))------- ( -35.90) >DroAna_CAF1 87849 116 + 1 GUACAAGAACAAGUUUCAAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGAACACAA----GAACCGAGGCGAAAGUCUGAGCUCUGGCGAGGAUCCCUGAUCCC ((...(((.(..((((((.((.....(((((((..((..(...)..))..)))).)))..)).)))).))..----......((((....))))..).))).))..(((((...))))). ( -30.10) >consensus GCAGGAGAACAAGUUUCGAGUGGAAAUGCGUGUAGCGUGCAGCGUGCGAAACACUGCAGAACAUGGACACGAAUCGAGAUCGAGGCGAAAGCCUGA_CGCUGAUGCAGAUCUC_______ ............((.((..(((....(((((((..((..(...)..))..)))).)))...))).)).)).....((((((.((((....))))....((....)).))))))....... (-31.70 = -31.57 + -0.14)


| Location | 18,974,232 – 18,974,337 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -47.26 |
| Consensus MFE | -33.44 |
| Energy contribution | -34.72 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18974232 105 + 27905053 CGAGGCGAAAGCCUGACGCUGAUGCAGAUCUCCUCC----CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-GGCCAUGAUGAUGGUGGCGCGGGUCGGAGG ..((((....))))...((....))....((((..(----((((((..((.(..(....((((..(((....)))...)-)))....)..).))..)))))))..)))). ( -48.20) >DroSec_CAF1 85287 105 + 1 CGAGGCGAAAGCCUGACGCUGAUGCAGAUCUCCUCC----CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-GGCAAUGGUGGUGGUGGCGCGGGUCGGAGG ..((((....))))...((....))....((((..(----((((((..((.(..(...(((((..(((....)))...)-))))...)..).))..)))))))..)))). ( -49.90) >DroSim_CAF1 87080 105 + 1 CGAGGCGAAAGCCUGACGCUGAUGCAGAUCUCCUCC----CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-GGCAAUGGUGGUGGCGGCGCGGGUCGGGGG ..((((....))))...((....))....((((..(----((((((((((.(..(...(((((..(((....)))...)-))))...)..).)))))))))))..)))). ( -51.50) >DroEre_CAF1 85884 101 + 1 CGAGGCGAAAGCCUGACGCUGAUGCAGAUCUCCUCC----CCGGGCCGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGGGUGGAAUGGUGG---CGGCGGGU--CGGUGG ..((((....))))..(((((((.(......((...----..))((((((((........(((((((((......)))))))))..)).))---))))).))--))))). ( -44.50) >DroYak_CAF1 88447 106 + 1 CGAGGCGAAAGCCUGACGCUGAUGCAGAUCUCCUCCUCCGCCGGGCCGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-UUUAAUGGUGG---CGGCGGGUUUUGGAGG ..((((....))))...((....))......(((((((((((..((((((....((((...(((.(((....))).)))-))))..)))))---)))))))....))))) ( -42.20) >consensus CGAGGCGAAAGCCUGACGCUGAUGCAGAUCUCCUCC____CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG_GGCAAUGGUGGUGGCGGCGCGGGUCGGAGG ..((((....))))...((....))......(((((....((((((((((.(..(......(((.(((....))).)))........)..).))))))))))...))))) (-33.44 = -34.72 + 1.28)

| Location | 18,974,264 – 18,974,375 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.88 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18974264 111 + 27905053 CUCC----CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-GGCCAUGAUGAUGGUGGCGCGGGUCGGAGGCAUGAAU-UUGCCAGCAUGAAAAAUGGCAACACUUUUGC ((((----((((((..((.(..(....((((..(((....)))...)-)))....)..).))..))))))...))))(((.....-((((((..........))))))......))) ( -39.10) >DroSec_CAF1 85319 111 + 1 CUCC----CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-GGCAAUGGUGGUGGUGGCGCGGGUCGGAGGCAUGGAU-UUGCCAGCAUGAAAAAUGGCAACACUUUUGC ((((----((((((..((.(..(...(((((..(((....)))...)-))))...)..).))..))))))...))))(((.((..-((((((..........))))))..))..))) ( -41.60) >DroSim_CAF1 87112 111 + 1 CUCC----CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-GGCAAUGGUGGUGGCGGCGCGGGUCGGGGGCAUGGAU-UUGCCAGCAUGAAAAAUGGCAACACUUUUGC ((((----((((((((((.(..(...(((((..(((....)))...)-))))...)..).)))))))))))..)))((((.....-.))))......((((.((....)).)))).. ( -43.90) >DroEre_CAF1 85916 107 + 1 CUCC----CCGGGCCGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGGGUGGAAUGGUGG---CGGCGGGU--CGGUGGCAUGGAU-UUGCCAGCAUGAAAAAUGGCAACACUUUUGC ....----((((.(((((.(..(.....(((((((((......)))))))))...)..)---.))))).)--)))(((((.....-.))))).....((((.((....)).)))).. ( -40.10) >DroYak_CAF1 88479 113 + 1 CUCCUCCGCCGGGCCGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG-UUUAAUGGUGG---CGGCGGGUUUUGGAGGCAUAGAUUUUGCCAGCAUGAAAAAUGGCAACACUUUUGC ((((((((((..((((((....((((...(((.(((....))).)))-))))..)))))---)))))))....))))(((......((((((..........))))))......))) ( -37.80) >consensus CUCC____CCGUGCUGCCCUUGUAAAUGUCCAUCUAAAAAUAGUUGG_GGCAAUGGUGGUGGCGGCGCGGGUCGGAGGCAUGGAU_UUGCCAGCAUGAAAAAUGGCAACACUUUUGC ((((....((((((((((.(..(......(((.(((....))).)))........)..).))))))))))...))))(((......((((((..........))))))......))) (-29.60 = -30.88 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:13 2006