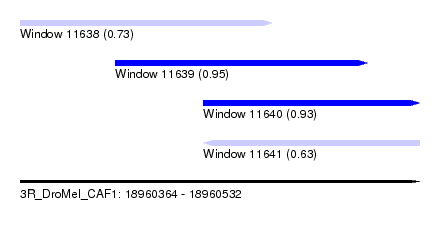
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,960,364 – 18,960,532 |
| Length | 168 |
| Max. P | 0.952735 |

| Location | 18,960,364 – 18,960,470 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -24.56 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18960364 106 + 27905053 CGAUGUUUACCCAACAUGCUAAUGCAUUGAGCUCGAAUUCGAAUCGAUGC--GCUGGACGCGCAAUGAACAAACAACGGCCCGUCUCAGAUCUCGGA----UCUGAG-------AUCUG ..(((((.....)))))......(((((((..(((....))).)))))))--...(((((.((..((......))...)).)))))(((((((((..----..))))-------))))) ( -33.30) >DroSec_CAF1 71778 106 + 1 CGAUGUUUACCCAACAUGCUAAUGCAUUGAGCUCGAAUUCGAAUCGAUGC--GCUGGACGCGCAAUGUACAAACAACGGCCCGUCUCAGAUCUCGGA----UCUGGG-------AACUG ((.(((((.....((((((....(((((((..(((....))).)))))))--((.....)))).))))..))))).)).....(((((((((...))----))))))-------).... ( -32.30) >DroSim_CAF1 73381 113 + 1 CGAUGUUUACCCAACAUGCUAAUGCAUUGAGCUCGAAUUCGAAUCGAUGC--GCUGGACGCGCAAUGUACAAACAACGGCCCGUCUCAGAUCUCGGA----UCUGGGAUCUGGGAUCUG ..(((((.....)))))......(((((((..(((....))).)))))))--...(((((.((..(((....)))...)).)))))(((((((((((----((...))))))))))))) ( -40.60) >DroEre_CAF1 71949 91 + 1 CGAUGUUUACCCAACAUGCUAAUGCAUUGAGCUCGAAUUCGAAUCGAUGC--GCUGGACGCGCAAUGAACAAACAACGGC-CGUCUCAGAU-------------------------CUC ..(((((.....)))))......(((((((..(((....))).)))))))--.(((((((.((..((......))...))-)))).)))..-------------------------... ( -24.80) >DroYak_CAF1 74172 111 + 1 CGAUGUUUACCCAACAUGCUAAUGCAUUGAGCUCGAAUUCGAAUCGAUGCGCGCUGGACGCGCAAUGUACAAACAACGGG-CGUCUCAGAUCUCAGAGAUCUCAGAG-------AUCUC .((((....(((...........(((((((..(((....))).)))))))((((.....))))..............)))-))))..(((((((.((....)).)))-------)))). ( -36.00) >consensus CGAUGUUUACCCAACAUGCUAAUGCAUUGAGCUCGAAUUCGAAUCGAUGC__GCUGGACGCGCAAUGUACAAACAACGGCCCGUCUCAGAUCUCGGA____UCUGAG_______AUCUG ..(((((.....)))))......(((((((..(((....))).)))))))...(((((((.((..((......))...)).)))).)))..((((((....))))))............ (-24.56 = -25.10 + 0.54)



| Location | 18,960,404 – 18,960,510 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -42.84 |
| Consensus MFE | -25.00 |
| Energy contribution | -27.93 |
| Covariance contribution | 2.93 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18960404 106 + 27905053 GAAUCGAUGC--GCUGGACGCGCAAUGAACAAACAACGGCCCGUCUCAGAUCUCGGA----UCUGAG-------AUCUGGGAUCUGAGUUCUUUGGGAUCUCUCGAGACUGGGAUCUCU ((((((.(((--((.....))))).)))................(((((((((((((----((...)-------)))))))))))))))))...((((((((........)))))))). ( -43.30) >DroSec_CAF1 71818 104 + 1 GAAUCGAUGC--GCUGGACGCGCAAUGUACAAACAACGGCCCGUCUCAGAUCUCGGA----UCUGGG-------AACUGGGAACUGGGUUC--GGGGAUCUCUCGAGACUGGGAUCUCG (((((..(((--((.....)))))............((((((.(((((((((...))----))))))-------)...)))..))))))))--.((((((((........)))))))). ( -41.10) >DroSim_CAF1 73421 111 + 1 GAAUCGAUGC--GCUGGACGCGCAAUGUACAAACAACGGCCCGUCUCAGAUCUCGGA----UCUGGGAUCUGGGAUCUGGGAACUGGGUUC--GGGGAUCUCUCGAGACUGGGAUCUCG .......(((--((.....)))))............((((((.((((((((((((((----((...))))))))))))))))...))).))--)((((((((........)))))))). ( -51.00) >DroEre_CAF1 71989 87 + 1 GAAUCGAUGC--GCUGGACGCGCAAUGAACAAACAACGGC-CGUCUCAGAU-------------------------CUCGGAUCUGGGUUC--GGGGAUC--UCGAAACUGGGAUCUCU ...(((.(((--((.....))))).)))...........(-((.(((((((-------------------------(...))))))))..)--))(((((--(((....)))))))).. ( -31.70) >DroYak_CAF1 74212 107 + 1 GAAUCGAUGCGCGCUGGACGCGCAAUGUACAAACAACGGG-CGUCUCAGAUCUCAGAGAUCUCAGAG-------AUCUCGGAUCUGGGUUC--GGGGAUC--UCGAAACUGGGAUCUCU ((((((((((((((.....))))..(((....)))....)-)))).(((((((..((((((.....)-------)))))))))))))))))--(((((((--(((....)))))))))) ( -47.10) >consensus GAAUCGAUGC__GCUGGACGCGCAAUGUACAAACAACGGCCCGUCUCAGAUCUCGGA____UCUGAG_______AUCUGGGAUCUGGGUUC__GGGGAUCUCUCGAGACUGGGAUCUCU (((((..........(((((.((..((......))...)).)))))(((((((((((..................))))))))))))))))...((((((((........)))))))). (-25.00 = -27.93 + 2.93)



| Location | 18,960,441 – 18,960,532 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -31.54 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18960441 91 + 27905053 CCCGUCUCAGAUCUCGGAUCUGAG-------AUCUGGGAUCUGAGUUCUUUGGGAUCUCUCGAGACUGGGAUCUCUCCUCGCAGAUCCCAUAUUCCAC .....(((((((((((((((...)-------)))))))))))))).....((((((((..((((...(((....))))))).))))))))........ ( -40.40) >DroSec_CAF1 71855 89 + 1 CCCGUCUCAGAUCUCGGAUCUGGG-------AACUGGGAACUGGGUUC--GGGGAUCUCUCGAGACUGGGAUCUCGCCUCGCAGAUCCCAUAUUCCAC (((.(((((((((...))))))))-------)...)))...(((((((--(((.....))))))..((((((((.((...))))))))))...)))). ( -36.50) >DroSim_CAF1 73458 96 + 1 CCCGUCUCAGAUCUCGGAUCUGGGAUCUGGGAUCUGGGAACUGGGUUC--GGGGAUCUCUCGAGACUGGGAUCUCGCCUCGCAGAUCCCAUAUUCCAC (((.((((((((((((((((...))))))))))))))))...)))(((--(((.....))))))..((((((((.((...))))))))))........ ( -46.30) >consensus CCCGUCUCAGAUCUCGGAUCUGGG_______AUCUGGGAACUGGGUUC__GGGGAUCUCUCGAGACUGGGAUCUCGCCUCGCAGAUCCCAUAUUCCAC ...(((((.((.(((((.(((.((.........)).))).))))).))...(((....))))))))(((((((((.....).))))))))........ (-31.54 = -31.10 + -0.44)


| Location | 18,960,441 – 18,960,532 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -26.63 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18960441 91 - 27905053 GUGGAAUAUGGGAUCUGCGAGGAGAGAUCCCAGUCUCGAGAGAUCCCAAAGAACUCAGAUCCCAGAU-------CUCAGAUCCGAGAUCUGAGACGGG ........((((((((.(((((...(....)..)))))..)))))))).....((((((((.(.(((-------(...)))).).))))))))..... ( -33.80) >DroSec_CAF1 71855 89 - 1 GUGGAAUAUGGGAUCUGCGAGGCGAGAUCCCAGUCUCGAGAGAUCCCC--GAACCCAGUUCCCAGUU-------CCCAGAUCCGAGAUCUGAGACGGG .(((.....(((((((.((((((.........))))))..))))))).--....)))...(((.(((-------..((((((...)))))).)))))) ( -33.80) >DroSim_CAF1 73458 96 - 1 GUGGAAUAUGGGAUCUGCGAGGCGAGAUCCCAGUCUCGAGAGAUCCCC--GAACCCAGUUCCCAGAUCCCAGAUCCCAGAUCCGAGAUCUGAGACGGG .........(((((((.((((((.........))))))..))))))).--...(((.(((..((((((.(.((((...)))).).)))))).)))))) ( -36.80) >consensus GUGGAAUAUGGGAUCUGCGAGGCGAGAUCCCAGUCUCGAGAGAUCCCC__GAACCCAGUUCCCAGAU_______CCCAGAUCCGAGAUCUGAGACGGG .(((.....(((((((.((((((.........))))))..)))))))...((((...)))))))............((((((...))))))....... (-26.63 = -27.30 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:48 2006