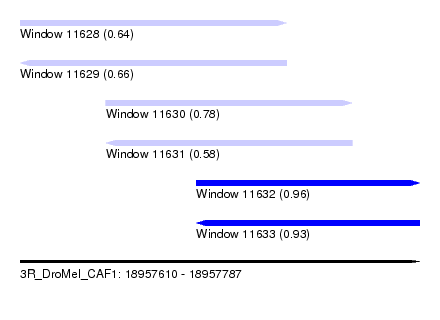
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,957,610 – 18,957,787 |
| Length | 177 |
| Max. P | 0.958496 |

| Location | 18,957,610 – 18,957,728 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18957610 118 + 27905053 AAAAAAAACAUGAACAUGAAGAAAAUGAAGUAUGAAAAAGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAA ........((((..(((.......)))...))))..................((((.....))))(((((..........)))))((((((((..(..(....)..).)))))))).. ( -24.40) >DroSec_CAF1 69039 105 + 1 -------------ACAUGAAGGAAAUGAAGUAUGAAAGAGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAA -------------.....(((..(((.((((........(....)(..((((.........))))..)......)))).)))..)))((((((..(..(....)..).)))))).... ( -22.40) >DroSim_CAF1 70185 105 + 1 -------------ACAUGAAGGAAAUGAAGUAUGAAAGAGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAA -------------.....(((..(((.((((........(....)(..((((.........))))..)......)))).)))..)))((((((..(..(....)..).)))))).... ( -22.40) >consensus _____________ACAUGAAGGAAAUGAAGUAUGAAAGAGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAA ..................((((.(((.((((........(....)(..((((.........))))..)......)))).))).))))((((((..(..(....)..).)))))).... (-22.82 = -22.60 + -0.22)

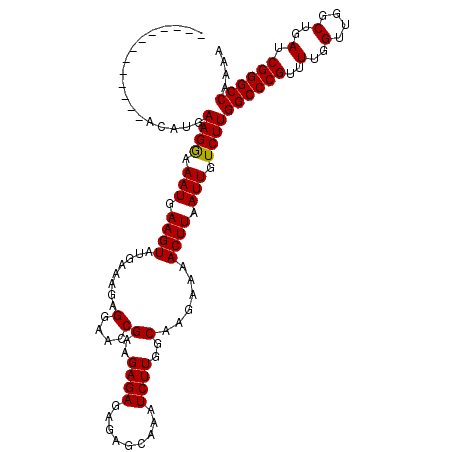
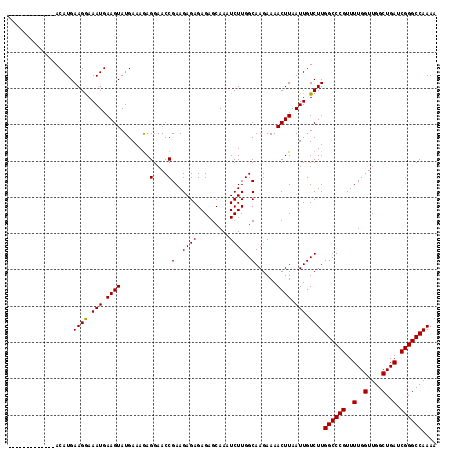
| Location | 18,957,610 – 18,957,728 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18957610 118 - 27905053 UUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCUUUUUCAUACUUCAUUUUCUUCAUGUUCAUGUUUUUUUU ..((((((((...............))))))))((.(((.((((......(((((((...........)))).)))...........)))).))).)).................... ( -18.59) >DroSec_CAF1 69039 105 - 1 UUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCUCUUUCAUACUUCAUUUCCUUCAUGU------------- ((((((((((...............)))))))))).(((.((((......(((((((...........)))).)))...........)))).)))..........------------- ( -17.79) >DroSim_CAF1 70185 105 - 1 UUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCUCUUUCAUACUUCAUUUCCUUCAUGU------------- ((((((((((...............)))))))))).(((.((((......(((((((...........)))).)))...........)))).)))..........------------- ( -17.79) >consensus UUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCUCUUUCAUACUUCAUUUCCUUCAUGU_____________ ((((((((((...............)))))))))).(((.((((......(((((((...........)))).)))...........)))).)))....................... (-17.79 = -17.79 + -0.00)

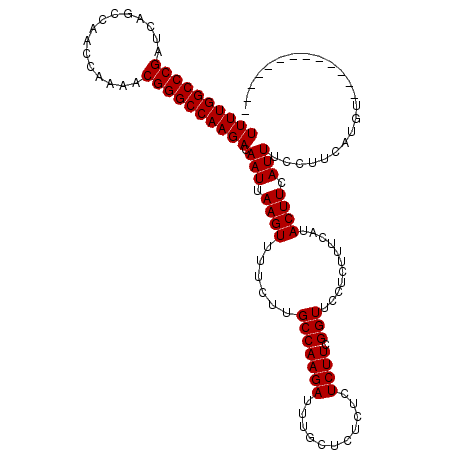
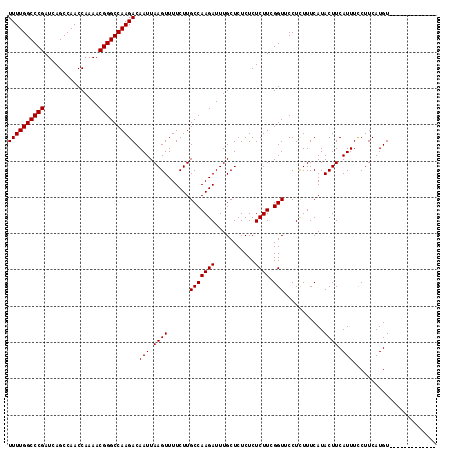
| Location | 18,957,648 – 18,957,757 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.42 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -24.71 |
| Energy contribution | -26.23 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18957648 109 + 27905053 AGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUC-----------GGUCUUUGGGGCAGUU ..(..(((((((..(((((.((((((((((((..........)))))((((((((..(..(....)..).))))))))))).))))..)))))-----------..)))))))..).... ( -39.20) >DroSec_CAF1 69064 109 + 1 AGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUC-----------GGUCUUUGGGGCAGUU ..(..(((((((..(((((.((((((((((((..........)))))((((((((..(..(....)..).))))))))))).))))..)))))-----------..)))))))..).... ( -39.20) >DroSim_CAF1 70210 109 + 1 AGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUC-----------GGUCUUUGGGGCAGUU ..(..(((((((..(((((.((((((((((((..........)))))((((((((..(..(....)..).))))))))))).))))..)))))-----------..)))))))..).... ( -39.20) >DroEre_CAF1 69220 109 + 1 AAGAACCAAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUC-----------GGUCUUUGGGGCAGUU ..(..(((((((..(((((.((((((((((((..........)))))((((((((..(..(....)..).))))))))))).))))..)))))-----------..)))))))..).... ( -39.70) >DroAna_CAF1 71268 106 + 1 -----------AAAG-AAGGAAAUCUUGGCGAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGGUCGUAU--GGAGCAUUUUCCUCUCUCUUGUUAUUUGGUGUUUGGGGCAGUU -----------.(((-(......))))(((((..........)))))(((.((((..(..(.((((.((......--((((....)))).....)).)))).)..)....)))).))).. ( -18.70) >consensus AGGAACCGAAGAGAGAGAGCAAAUCUUGGCAAGAAAACUUAAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUC___________GGUCUUUGGGGCAGUU ..(..((((((((((.(((.((((((((((((..........)))))((((((((..(..(....)..).))))))))))).)))).)))))).............)))))))..).... (-24.71 = -26.23 + 1.52)

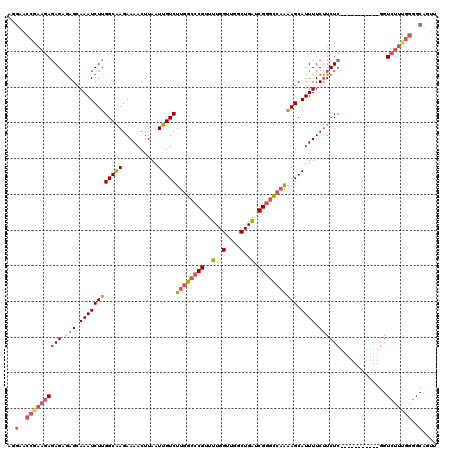
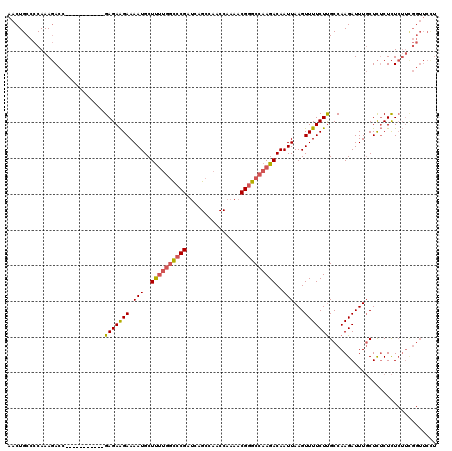
| Location | 18,957,648 – 18,957,757 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.42 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18957648 109 - 27905053 AACUGCCCCAAAGACC-----------GAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCU ............((((-----------((..(((....((((((((((((...............)))))))))).....(((((((......)))))))))....)))..))))))... ( -28.46) >DroSec_CAF1 69064 109 - 1 AACUGCCCCAAAGACC-----------GAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCU ............((((-----------((..(((....((((((((((((...............)))))))))).....(((((((......)))))))))....)))..))))))... ( -28.46) >DroSim_CAF1 70210 109 - 1 AACUGCCCCAAAGACC-----------GAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCU ............((((-----------((..(((....((((((((((((...............)))))))))).....(((((((......)))))))))....)))..))))))... ( -28.46) >DroEre_CAF1 69220 109 - 1 AACUGCCCCAAAGACC-----------GAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUUGGUUCUU ....(..(((((((..-----------(((((((((((..((((((((((...............)))))))))).......))))))).((........))))))..)))))))..).. ( -31.66) >DroAna_CAF1 71268 106 - 1 AACUGCCCCAAACACCAAAUAACAAGAGAGAGGAAAAUGCUCC--AUACGACCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUCGCCAAGAUUUCCUU-CUUU----------- ........................(((((.((((((..(((..--........)))...........(((.(((((......)))))...)))....)))))))-))))----------- ( -14.90) >consensus AACUGCCCCAAAGACC___________GAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUUAAGUUUUCUUGCCAAGAUUUGCUCUCUCUCUUCGGUUCCU ...........................(((((((.(((..((((((((((...............))))))))))..)))...))))))).............................. (-17.12 = -17.48 + 0.36)


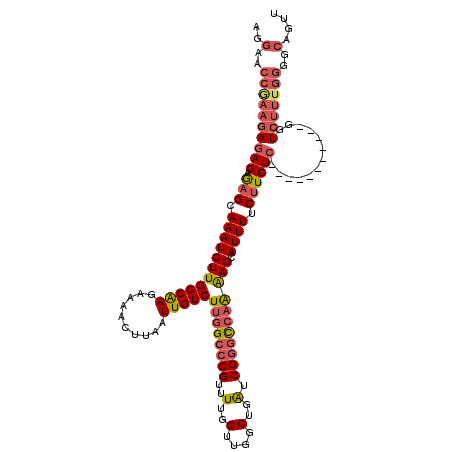
| Location | 18,957,688 – 18,957,787 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 99.19 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18957688 99 + 27905053 AAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUCGGUCUUUGGGGCAGUUUCGUUAUGUUUCGGGGAAAACUGGUUGCUG .......((((((((..(..(....)..).)))))))).((((....((......((((((..(((.........)))..)))))).....)).)))). ( -31.80) >DroSec_CAF1 69104 99 + 1 AAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUCGGUCUUUGGGGCAGUUUCGUUAUGUUUCGGGGAAAACUGGUUGCUG .......((((((((..(..(....)..).)))))))).((((....((......((((((..(((.........)))..)))))).....)).)))). ( -31.80) >DroSim_CAF1 70250 99 + 1 AAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUCGGUCUUUGGGGCAGUUUCGUUAUGUUUCGGGGAAAACUGGUUGCUG .......((((((((..(..(....)..).)))))))).((((....((......((((((..(((.........)))..)))))).....)).)))). ( -31.80) >DroEre_CAF1 69260 99 + 1 AAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUCGGUCUUUGGGGCAGUUUCGUUAUGUUUCGGGGAAAACUGGUUGCUG .......((((((((..(..(....)..).)))))))).((((....((......((((((..(((.........)))..)))))).....)).)))). ( -31.80) >DroYak_CAF1 71376 99 + 1 AAUUGUCUUGGCCCGUUUUGGUUGGCUGAACGGGCCAAAAGCAUUUUCUUCUCGGCCUUUGGGGCAGUUUCGUUAUGUUUCGGGGAAAACUGGUUGCUG .......(((((((((.(..(....)..)))))))))).((((....((......((((.(..(((.........)))..)))))......)).)))). ( -30.80) >consensus AAUUGUCUUGGCCCGUUUUGGUUGGCUGAUCGGGCCAAAAGCAUUUUCUUCUCGGUCUUUGGGGCAGUUUCGUUAUGUUUCGGGGAAAACUGGUUGCUG .......((((((((..(..(....)..).)))))))).((((....((......((((((..(((.........)))..)))))).....)).)))). (-30.30 = -30.50 + 0.20)

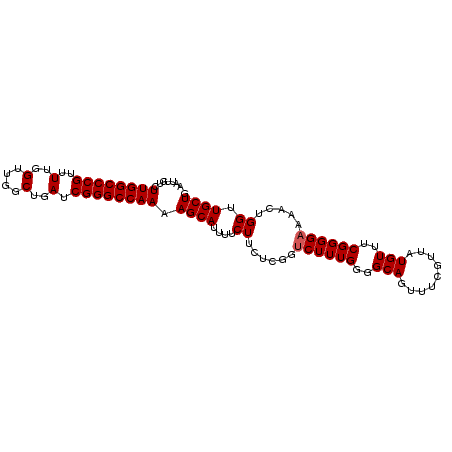

| Location | 18,957,688 – 18,957,787 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 99.19 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18957688 99 - 27905053 CAGCAACCAGUUUUCCCCGAAACAUAACGAAACUGCCCCAAAGACCGAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUU .......(((((((..............))))))).....................((.((((((((((...............))))))))))))... ( -19.30) >DroSec_CAF1 69104 99 - 1 CAGCAACCAGUUUUCCCCGAAACAUAACGAAACUGCCCCAAAGACCGAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUU .......(((((((..............))))))).....................((.((((((((((...............))))))))))))... ( -19.30) >DroSim_CAF1 70250 99 - 1 CAGCAACCAGUUUUCCCCGAAACAUAACGAAACUGCCCCAAAGACCGAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUU .......(((((((..............))))))).....................((.((((((((((...............))))))))))))... ( -19.30) >DroEre_CAF1 69260 99 - 1 CAGCAACCAGUUUUCCCCGAAACAUAACGAAACUGCCCCAAAGACCGAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUU .......(((((((..............))))))).....................((.((((((((((...............))))))))))))... ( -19.30) >DroYak_CAF1 71376 99 - 1 CAGCAACCAGUUUUCCCCGAAACAUAACGAAACUGCCCCAAAGGCCGAGAAGAAAAUGCUUUUGGCCCGUUCAGCCAACCAAAACGGGCCAAGACAAUU ..(((....(((((....))))).........(.(((.....))).).........)))((((((((((((...........))))))))))))..... ( -25.30) >consensus CAGCAACCAGUUUUCCCCGAAACAUAACGAAACUGCCCCAAAGACCGAGAAGAAAAUGCUUUUGGCCCGAUCAGCCAACCAAAACGGGCCAAGACAAUU .......(((((((..............))))))).....................((.((((((((((...............))))))))))))... (-19.46 = -19.46 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:41 2006