
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,955,764 – 18,956,005 |
| Length | 241 |
| Max. P | 0.975560 |

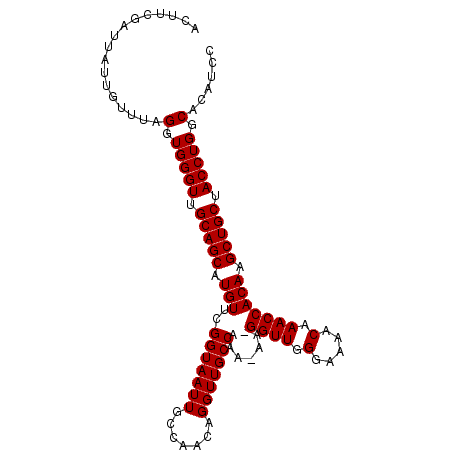
| Location | 18,955,764 – 18,955,867 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 98.64 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18955764 103 - 27905053 ACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AAUGGUUGGGAAAACAAACCACAAGCUGCUACCUGGCACAUCC .................(.(((((.(((((.(((..(((((((.......)))))))...-...((((.(.....).))))))).))))).))))).)...... ( -28.90) >DroSec_CAF1 67239 102 - 1 ACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AA-GGUUGGGAAAACAAACCACAAGCUGCUACCUGGCACAUCC .................(.(((((.(((((.(((..(((((((.......)))))))...-..-((((.(.....).))))))).))))).))))).)...... ( -29.10) >DroSim_CAF1 68392 102 - 1 ACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AA-GGUUGGGAAAACAAACCACAAGCUGCUACCUGGCACAUCC .................(.(((((.(((((.(((..(((((((.......)))))))...-..-((((.(.....).))))))).))))).))))).)...... ( -29.10) >DroEre_CAF1 67382 103 - 1 ACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAAAAA-GGUUGGGAAAACAAACCACAAGCUGCUACCUGGCCCAUCC .................(((((((((((((.(((..(((((((.......)))))))......-((((.(.....).))))))).))))).....)))))))). ( -31.10) >DroYak_CAF1 69408 102 - 1 ACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AA-GGUUGGGAAAACAAACCACAAGCUGCUACCUGGCCCAUCC .................(((((((((((((.(((..(((((((.......)))))))...-..-((((.(.....).))))))).))))).....)))))))). ( -31.10) >consensus ACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA_AA_GGUUGGGAAAACAAACCACAAGCUGCUACCUGGCACAUCC .................(.(((((.(((((.(((..(((((((.......))))))).......((((.(.....).))))))).))))).))))).)...... (-28.54 = -28.54 + -0.00)

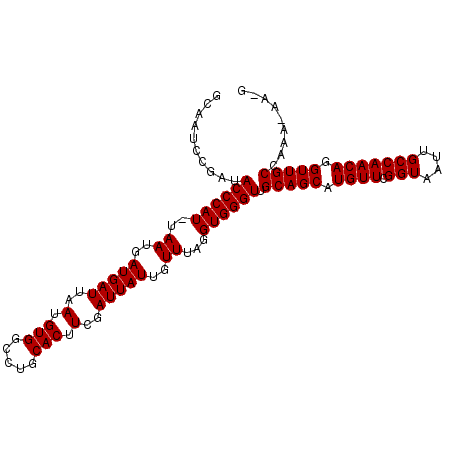
| Location | 18,955,803 – 18,955,906 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -29.56 |
| Energy contribution | -29.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18955803 103 - 27905053 GCAAUCCGAUACCCAU-UAAUGAUGAUUAAUGUGGCCUGCACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AAUG ..........((((((-(((..(((((..(.(((.....))).)..)))))..))..))))))).(((((.((((.(((....))))))).)))))....-.... ( -28.70) >DroSec_CAF1 67278 102 - 1 GCAAUCCGAUACCCAU-UAAUGAUGAUUAAUGUGGCCUGCACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AA-G ..........((((((-(((..(((((..(.(((.....))).)..)))))..))..))))))).(((((.((((.(((....))))))).)))))....-..-. ( -28.70) >DroSim_CAF1 68431 102 - 1 GCAAUCCGAUACCCAU-UAAUGAUGAUUAAUGUGGCCUGCACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AA-G ..........((((((-(((..(((((..(.(((.....))).)..)))))..))..))))))).(((((.((((.(((....))))))).)))))....-..-. ( -28.70) >DroEre_CAF1 67421 104 - 1 GCAAUCGGAUACCCAUUUAAUGAUGAUUAAUGUGGCCAGCACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAAAAA-G ..........((((((((((..(((((..(.(((.....))).)..)))))..).))))))))).(((((.((((.(((....))))))).))))).......-. ( -30.60) >DroYak_CAF1 69447 102 - 1 GCAAUCCGAUACCCAU-UAAUGAUGAUUAAUGUGGCCUGCACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA-AA-G ..........((((((-(((..(((((..(.(((.....))).)..)))))..))..))))))).(((((.((((.(((....))))))).)))))....-..-. ( -28.70) >consensus GCAAUCCGAUACCCAU_UAAUGAUGAUUAAUGUGGCCUGCACUUCGAUUAUUGUUUAGGUGGGUUGCAGCAUGUUCGGUAAUUGCCAACAGGUUGCCAAA_AA_G ..........((((((..((..(((((..(.(((.....))).)..)))))..))...)))))).(((((.((((.(((....))))))).)))))......... (-29.56 = -29.56 + 0.00)


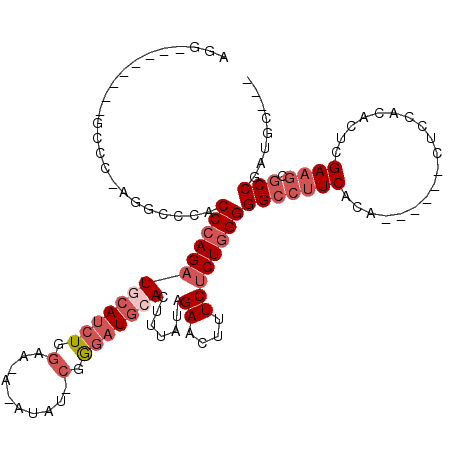
| Location | 18,955,906 – 18,956,005 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -35.99 |
| Consensus MFE | -16.65 |
| Energy contribution | -18.34 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18955906 99 + 27905053 AGG--------GAAC-AGGCCCACCCAGAUGCAUCUGGAA--UAUAU-CGAGAUGCACUCUAUAGAACUUUCUCUGGGGGCCUUCACA------CUCCACACUCGAAGCGCCGAUGC--- .((--------(...-...))).(((((((((((((.((.--....)-).))))))).((....))......))))))(((((((...------..........)))).))).....--- ( -33.52) >DroSec_CAF1 67380 101 + 1 AGG--------GCCC-AGGCCCACCCAGAUGCAUCUGGAACAUAUAU-CGGGAUGCACUUUAUAGAACUUUCUCUGGGGGCCUUCACA------CUCCACACUCGAAGCGCCGAUGC--- .((--------((..-..)))).(((((((((((((.((.......)-).)))))))((....)).......))))))(((((((...------..........)))).))).....--- ( -38.02) >DroSim_CAF1 68533 101 + 1 AGG--------GCCC-AGGCCCACCCAGAUGCAUCUGGAACAUAUAU-CGGGAUGCACUUUAUAGAACUUUCUCUGGGGGCCUUCACA------CUCCACACUCGAAGCGCCGAUGC--- .((--------((..-..)))).(((((((((((((.((.......)-).)))))))((....)).......))))))(((((((...------..........)))).))).....--- ( -38.02) >DroEre_CAF1 67525 93 + 1 AGG--------GCCAGGGGCCCACCCAGAUGCAUCUGGAGAA--UAU-CGGGAUGGACUUUGUAGAACUUUCUCUGGGGGCCUUCAC-------------ACUCGAAGCGCCGAUGC--- .((--------(((...))))).((((((..(((((.((...--..)-).)))))..((....)).......))))))(((((((..-------------....)))).))).....--- ( -36.50) >DroYak_CAF1 69549 102 + 1 AGGGACAAGGGGCCAGGGGCCCACCCAGAUGCAUCUGGAAAA--UGCACGGCAUGCACUUUGUAGAACUUUCUCUGGGGGCCUUCAC-------------ACUCGAAGCGCCGAUGC--- .........(((((...))))).((((((....((((.(((.--((((.....)))).))).))))......))))))(((((((..-------------....)))).))).....--- ( -36.90) >DroAna_CAF1 69616 105 + 1 AGG--------CAUC--GGGCCACCCAGCUAGAUCUGGAA---AUAU-AGGGCUGCAUUUU-CAGAACUUUCUCUUGGGGGGCUCACACGCACAUUUCACACCCGAAGCGCCGCCGCCGC .((--------(...--(((((.(((((..((.(((((((---((.(-(....)).)))))-)))).)).....))))).)))))...(((....(((......))))))..)))..... ( -33.00) >consensus AGG________GCCC_AGGCCCACCCAGAUGCAUCUGGAA_A_AUAU_CGGGAUGCACUUUAUAGAACUUUCUCUGGGGGCCUUCACA______CUCCACACUCGAAGCGCCGAUGC___ .......................(((((((((((((.(..........).))))))).......((....))))))))(((((((...................)))).)))........ (-16.65 = -18.34 + 1.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:33 2006