
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,955,327 – 18,955,471 |
| Length | 144 |
| Max. P | 0.894263 |

| Location | 18,955,327 – 18,955,440 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.34 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -31.49 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18955327 113 + 27905053 -ACUGAGAGCCGCCCAGGUAAUGGCAUUAGUUUGCUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGU -..((((((((((....))..(((((((((....))))).....((((.(((..(((......)))..)))))))...))))....)))))))).....------.((((.....)))). ( -30.50) >DroSec_CAF1 66805 113 + 1 -ACUGAGGGCCGCCCAGGUAAUGGCAUUAGUUUGUUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGU -.....(((((((((......(((((((((.........)))))((((.(((..(((......)))..)))))))...))))....((.((.....)).------)).)))).))))).. ( -34.20) >DroSim_CAF1 67958 113 + 1 -ACUGAGGGCCGCCCAGGUAAUGGCAUUAGUUUGCUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGU -.....(((((((((.((...(((((......)))))((((...((((.(((..(((......)))..)))))))...((.(((.....))).))))))------)).)))).))))).. ( -36.40) >DroEre_CAF1 66944 113 + 1 -GCUGAGGGCCGCCCAGGUAAUGGCAUUAGUUUGCUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGU -.....(((((((((.((...(((((......)))))((((...((((.(((..(((......)))..)))))))...((.(((.....))).))))))------)).)))).))))).. ( -36.40) >DroYak_CAF1 68940 113 + 1 -ACUGAGGGCCGCCCAGGUAAUGGCAUUAGUUUGCUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGU -.....(((((((((.((...(((((......)))))((((...((((.(((..(((......)))..)))))))...((.(((.....))).))))))------)).)))).))))).. ( -36.40) >DroAna_CAF1 69021 119 + 1 GGGUGUAGUUGGCCCAGGUAAUAGCAUUAGU-UGCUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAACCCAACAAGUCAAAAAGGUUUUCACAGAUCGAUCCCCGGGGCUGGCCCGU ((((((..(((((((((((..(((((.....-))))).....)))))).)))))))))))....((((..(((.............((((......))))(.....))))..)))).... ( -33.10) >consensus _ACUGAGGGCCGCCCAGGUAAUGGCAUUAGUUUGCUAAUCUAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU______CCGGGGCUGGCCCGU ......(((((((((.((...(((((......)))))((((...((((.(((..(((......)))..)))))))...((.(((.....))).))))))......)).)))).))))).. (-31.49 = -31.60 + 0.11)



| Location | 18,955,366 – 18,955,471 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -26.57 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18955366 105 + 27905053 UAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGUUGCAGCCCAAUGUGAAACAAUGGACUCG---------AAA ....((((.(((..(((......)))..)))))))(.((((......((((.((((...------...((((((........))))))..))))))))....)))).)---------... ( -30.50) >DroSec_CAF1 66844 105 + 1 UAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGUUGCAGCCCAAUGUGAAACAAUGGGCACG---------AAA ....((((((.............))))))(((((.....((.....)).(((((((...------...((((((........))))))..)))))))...)))))...---------... ( -31.62) >DroSim_CAF1 67997 105 + 1 UAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGUUGCAGCCCAAUGUGAAACAAUGGGCACG---------AAA ....((((((.............))))))(((((.....((.....)).(((((((...------...((((((........))))))..)))))))...)))))...---------... ( -31.62) >DroEre_CAF1 66983 105 + 1 UAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGUUGCAGCCCAAUGUGAAACAAUGGGCCCG---------AAA ....((((((.............))))))(((((.....((.....)).(((((((...------...((((((........))))))..)))))))...)))))...---------... ( -30.72) >DroYak_CAF1 68979 105 + 1 UAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU------CCGGGGCUGGCCCGUUGCAGCCCAAUGUGAAACAAUGGGCCCG---------AAA ....((((((.............))))))(((((.....((.....)).(((((((...------...((((((........))))))..)))))))...)))))...---------... ( -30.72) >DroAna_CAF1 69060 120 + 1 UAAUUUGGCGUUAAGCAUCCAAUUGCCAAACCCAACAAGUCAAAAAGGUUUUCACAGAUCGAUCCCCGGGGCUGGCCCGUCACAGCCCAAACUGAAACAAUGGGCCCCGAAAAAAAAAAA ...(((((((.............)))))))((((.....(((....((((......))))........((((((........))))))....))).....))))................ ( -27.72) >consensus UAAUUUGGCGUUAAGCAUCCAAUUGCCAAGCCCAACAAGUCAAAAAGGUUUUCACAGAU______CCGGGGCUGGCCCGUUGCAGCCCAAUGUGAAACAAUGGGCCCG_________AAA ....((((.(((..(((......)))..)))))))...(((......((((.((((............((((((........))))))..))))))))....)))............... (-26.57 = -26.62 + 0.06)

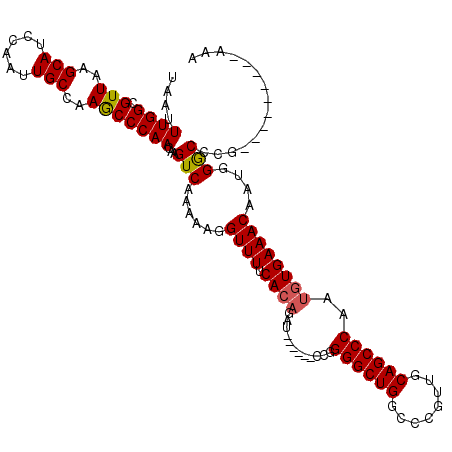
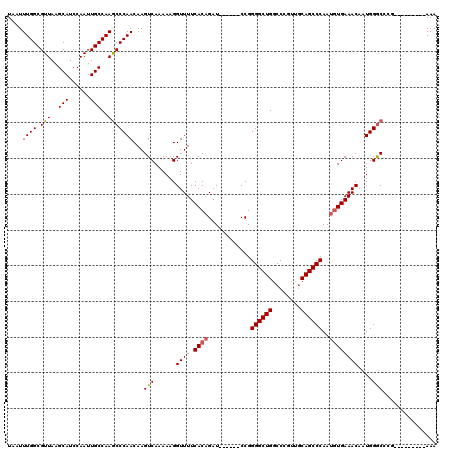
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:30 2006